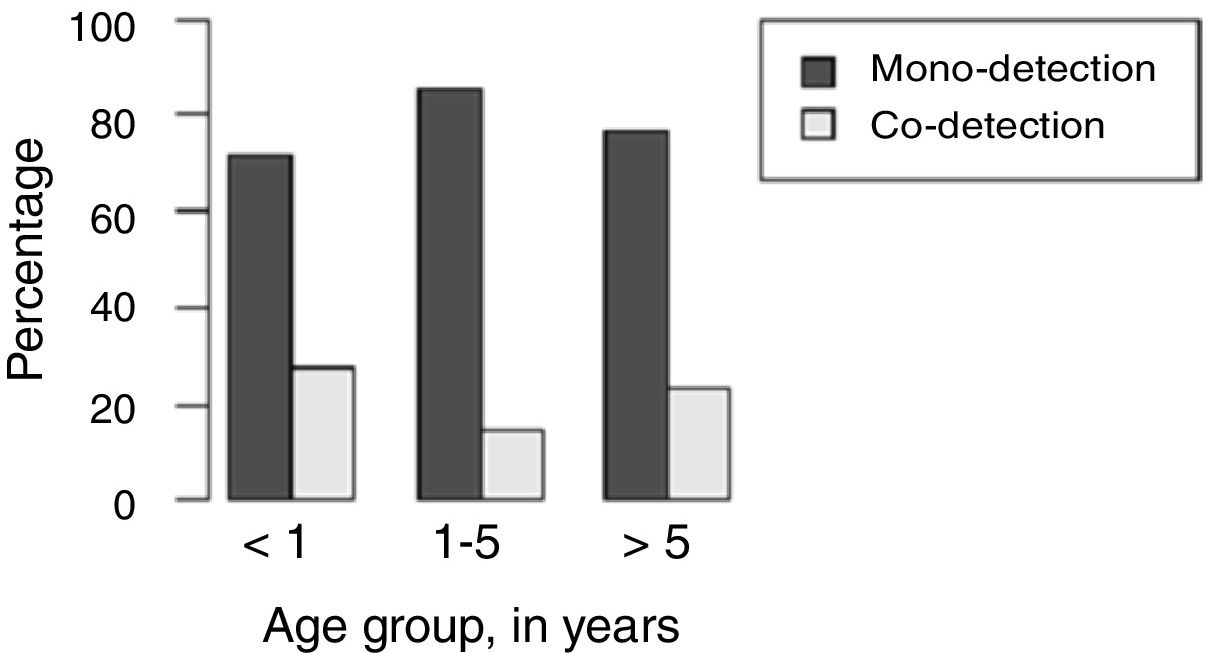The role of viral co-detection in children with severe acute respiratory infection is not clear. We described the viral detection profile and its association with clinical characteristics in children admitted to the Pediatric Intensive Care Unit (PICU) during the 2009 influenza A(H1N1) pandemic.
MethodLongitudinal observational retrospective study, with patients aged 0–18 years, admitted to 11 PICUs in Rio de Janeiro, with suspected H1N1 infection, from June to November, 2009. The results of respiratory samples which were sent to the Laboratory of Fiocruz/RJ and clinical data extracted from specific forms were analyzed.
ResultsOf 71 samples, 38% tested positive for H1N1 virus. Of the 63 samples tested for other viruses, 58 were positive: influenza H1N1 (43.1% of positive samples), rhinovirus/enterovirus (41.4%), respiratory syncytial vírus (12.1%), human metapneumovirus (12.1%), adenovirus (6.9%), and bocavirus (3.5%). Viral codetection occured in 22.4% of the cases. H1N1-positive patients were of a higher median age, had higher frequency of fever, cough and tachypnea, and decreased leukometry when compared to H1N1-negative patients. There was no difference in relation to severity outcomes (number of organic dysfunctions, use of mechanical ventilation or amines, hospital/PICU length of stay or death). Comparing the groups with mono-detection and co-dection of any virus, no difference was found regarding the association with any clinical variable.
ConclusionsOther viruses can be implicated in SARI in children. The role of viral codetection has not yet been completely elucidated.
Respiratory infections are an important cause of morbidity and mortality throughout the world, particularly in children.1–3 Besides the high susceptibility to viral respiratory infections, children present with a higher frequency of coinfections, especially those under five years of age.4,5
The influenza A (H1N1) pandemic in 2009, affected mainly children, young adults, pregnant women, and patients with chronic diseases. The overall severity of the disease was markedly increased compared to other influenza infections and there was a disproportionate increase in mortality.4 The hospitalization rate for influenza H1N1 in children in Argentina was twice the rate for seasonal influenza in the previous year.6 This pandemic quickly became a global health issue.
With the development of better laboratory methods for virus detection, the isolation of these agents in samples of respiratory secretions has become easier and more common than before. Studies evaluating viral codetection in children during the pandemic in Brazil and in other countries confirmed that other viruses, with similar cold weather seasonality,7 circulated concomitantly with H1N1.8 However, the relationship between the presence of other respiratory viruses and clinical severity in pediatric patients is still unclear. Some studies have described an association between viral coinfection with H1N1 and worse prognosis,9–11 yet others did not show any significant difference in outcomes.4,6,7,12 Thus, the primary objective of this study was to describe the viral detection profile and clinical characteristics of children admitted to the Pediatric Intensive Care Unit (PICU) with severe acute respiratory infection (SARI) during the 2009 epidemic in Rio de Janeiro. The association of viral detection with clinical characteristics at admission and resulting outcomes was explored as secondary objectives.
Material and methodsThis is a descriptive longitudinal observational study using retrospective data. A convenience sampling of patients ranging from 0 to 18 years of age, admitted to the PICU with severe acute respiratory syndrome (SARS), defined as fever, cough, and dyspnea, requiring hospitalization, during the 2009 pandemic in Rio de Janeiro, Brazil, according to the Brazilian Protocol of Clinical Management and Epidemiological Surveillance of Influenza13 was utilized.
SARI is the term most frequently used in the literature to describe severe cases, with a slightly different definition by the World Health Organization (WHO), as “an acute respiratory illness with a history of fever or measured fever of ≥38°C and cough, with onset within the past 10 days, requiring hospitalization”.14 In the present study, the term SARI will be used as synonymous of SARS for simplicity.
During this period, samples of respiratory secretions (nasopharyngeal swabs, nasopharyngeal aspirates or tracheal aspirates) of hospitalized patients with SARI were sent to three reference centers for influenza virus in Brazil. In the present study, we included 71 respiratory samples from patients who were admitted to the PICUs of six private hospitals and five public hospitals, from June 1 to November 30, 2009. These samples were tested for H1N1 virus in the respiratory virus laboratory of the reference center Oswaldo Cruz Foundation in Rio de Janeiro (Fiocruz/RJ), using the Real-time Polymerase Chain Reaction (RT-PCR) kit for H1N1 virus provided by the United States Centers for Disease Control and Prevention (CDC).
These samples, which had been stored at −70°C, were retrieved for further testing. Sixty-three samples were found to be viable and were retested for the presence of other infectious agents using one of the following three techniques, depending on availability at the Fiocruz laboratory: RT-PCR for non-influenza respiratory viruses from a CDC-based kit, RespiFinder (RespiFinder 22 kit; Pathofinder B.V., The Netherlands), or the Fast-track (FTDRP 21 plus multiplex real-time RT-PCR assay; Fast-track Diagnostics Ltd, Sliema, Malta).
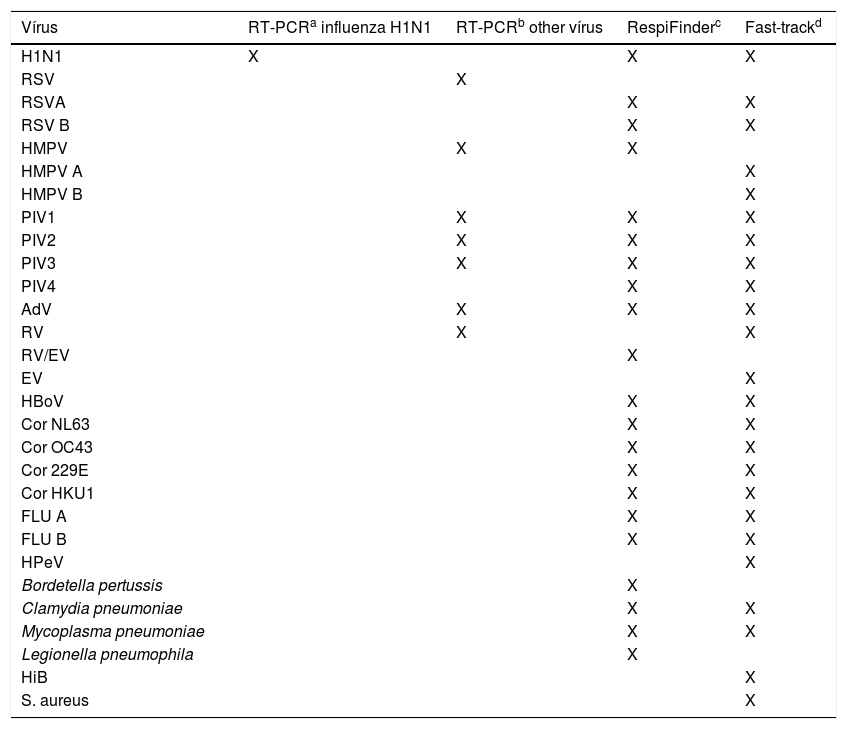
Seven respiratory viruses were screened by using RT-PCR for non-influenza respiratory viruses from a CDC-based kit; 17 respiratory viruses were screened by using the RespiFinder and 20 respiratory viruses and five bacteria were screened by using the Fast-track (Table 1). The RespiFinder and the Fast-track methods test a very similar virus profile. The differences are that the Fast-track identifies Rhinovirus and Enterovirus separately, discriminates Human Metapneumovirus in A and B, and test for Parechovirus, while the RespiFinder does not distinguish Rhinovirus from Enterovirus (reported as RV/EV), identifies Human Metapneumovirus in general and does not test for Parechovirus (Table 1).
Virus profile identified by four different laboratory methods.
| Vírus | RT-PCRa influenza H1N1 | RT-PCRb other vírus | RespiFinderc | Fast-trackd |
|---|---|---|---|---|
| H1N1 | X | X | X | |
| RSV | X | |||
| RSVA | X | X | ||
| RSV B | X | X | ||
| HMPV | X | X | ||
| HMPV A | X | |||
| HMPV B | X | |||
| PIV1 | X | X | X | |
| PIV2 | X | X | X | |
| PIV3 | X | X | X | |
| PIV4 | X | X | ||
| AdV | X | X | X | |
| RV | X | X | ||
| RV/EV | X | |||
| EV | X | |||
| HBoV | X | X | ||
| Cor NL63 | X | X | ||
| Cor OC43 | X | X | ||
| Cor 229E | X | X | ||
| Cor HKU1 | X | X | ||
| FLU A | X | X | ||
| FLU B | X | X | ||
| HPeV | X | |||
| Bordetella pertussis | X | |||
| Clamydia pneumoniae | X | X | ||
| Mycoplasma pneumoniae | X | X | ||
| Legionella pneumophila | X | |||
| HiB | X | |||
| S. aureus | X |
H1N1, influenza A(H1N1)pdm09; RV, rhinovirus; EV, enterovirus; RSV, respiratory syncytial virus; HMPV, human metapneumovirus; AdV, adenovirus; PIV, parainfluenza vírus; HBoV, human bocavirus; Flu, seasonal influenza vírus; Cor, coronavirus; HPeV, Parechovirus; HiB, Hemophilus influenza B; S aureus, Staphilococcus aureus.
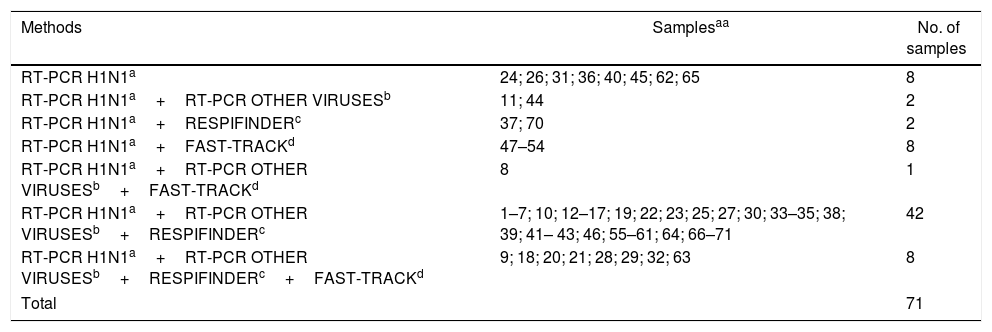
Eight samples were tested only with the RT-PCR kit for H1N1 virus during the epidemic, while 63 samples (89%) were retested later at least with two methods, including the Respifinder and/or the Fast track in 61 samples (86%) (Table 2).
Laboratory methods performed by samples from 71 different patients.
| Methods | Samplesaa | No. of samples |
|---|---|---|
| RT-PCR H1N1a | 24; 26; 31; 36; 40; 45; 62; 65 | 8 |
| RT-PCR H1N1a+RT-PCR OTHER VIRUSESb | 11; 44 | 2 |
| RT-PCR H1N1a+RESPIFINDERc | 37; 70 | 2 |
| RT-PCR H1N1a+FAST-TRACKd | 47–54 | 8 |
| RT-PCR H1N1a+RT-PCR OTHER VIRUSESb+FAST-TRACKd | 8 | 1 |
| RT-PCR H1N1a+RT-PCR OTHER VIRUSESb+RESPIFINDERc | 1–7; 10; 12–17; 19; 22; 23; 25; 27; 30; 33–35; 38; 39; 41– 43; 46; 55–61; 64; 66–71 | 42 |
| RT-PCR H1N1a+RT-PCR OTHER VIRUSESb+RESPIFINDERc+FAST-TRACKd | 9; 18; 20; 21; 28; 29; 32; 63 | 8 |
| Total | 71 | |
In order to evaluate the presence of H1N1, we considered a positive result from any of the four tests in the 71 samples. For the assessment of viral codetection, we considered the 63 samples which were retested by at least one of the three methods, other than the RT-PCR kit for H1N1 virus. We used the term codetection and not coinfection because it is difficult to attribute a causal relationship between the detected agents and the clinical picture of the patient.13
Demographic and clinical data were extracted from a specific data collection form for patients admitted to the PICU with SARI during the epidemic.
Continuous variables were described as median and interquartile range and categorical variables were described as proportions. To compare H1N1-positive versus H1N1-negative groups and viral mono-dectection and codetection groups we used age-adjusted logistic regressions for categorical variables and non-parametric ANCOVA for continuous variables. In case of categories with zero occurrences, we used the Mantel–Haenszel test to adjust for age.
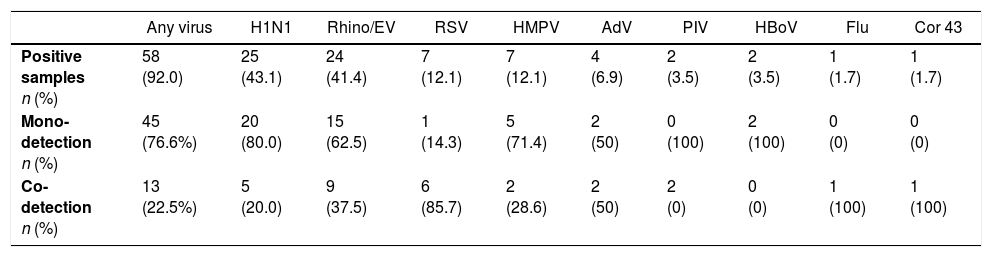
ResultsOf the 71 samples analyzed in 2009, 27 (38%) were positive for H1N1. Of the 63 samples retested in 2015, 58 (92.1%) were positive for one or more viruses. with the following descending order of frequency: influenza H1N1 (43.1% of positive samples), RV-EV (41.4%), RSV (12.1%), HMPV (12.1%), adenovirus (6.9%), bocavirus (3.5%) parainfluenza (3.5%), coronavirus 43 (1.7%), and seasonal influenza (1.7%). Of these 58 positive samples, 45 were mono-detections (77.6%) and 13 were codetections (22.5%), 11 with two viruses (19.0%), and two with three viruses (3.5%). The most frequent combination of codetected viruses were RV-EV and RSV (four of 13 samples with two or more viruses). Of the original 27 H1N1-positive samples, 25 were retested and five (20%) showed codetection with other viruses. The H1N1 virus was present in 46.5% of the samples with a single virus (20/43) and in 33.3% of the samples with codetection (5/13) (Table 3).
Mono-detection and co-detection of viruses in 63 respiratory samples from patients admitted to the PICU during the 2009 influenza A(H1N1) pandemic in Rio de Janeiro, Brazil.
| Any virus | H1N1 | Rhino/EV | RSV | HMPV | AdV | PIV | HBoV | Flu | Cor 43 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Positive samples n (%) | 58 (92.0) | 25 (43.1) | 24 (41.4) | 7 (12.1) | 7 (12.1) | 4 (6.9) | 2 (3.5) | 2 (3.5) | 1 (1.7) | 1 (1.7) |
| Mono-detection n (%) | 45 (76.6%) | 20 (80.0) | 15 (62.5) | 1 (14.3) | 5 (71.4) | 2 (50) | 0 (100) | 2 (100) | 0 (0) | 0 (0) |
| Co-detection n (%) | 13 (22.5%) | 5 (20.0) | 9 (37.5) | 6 (85.7) | 2 (28.6) | 2 (50) | 2 (0) | 0 (0) | 1 (100) | 1 (100) |
| CO-detection matrix | H1N1 | Rhino/EV | RSV | HMPV | AdV | PIV | HBoV | Flu | CoV 43 | |
|---|---|---|---|---|---|---|---|---|---|---|
| H1N1 n (%) | – | 1 (4.2) | 1 (14.3) | 1 (14.3) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | |
| RSV n (%) | 1 (4.0) | 4 (16.7) | – | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Rhino n (%) | 1 (4.0) | – | 4 (57.1) | 1 (14.3) | 0 (0) | 2 (100) | 0 (0) | 0 (0) | 0 (0) | |
| HMPV n (%) | 1 (4.0) | 1 (4.2) | 0 (0) | – | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Flu n (%) | 1 (4.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | – | 0 (0) | |
| PIV n (%) | 0 (0) | 2 (8.3) | 0 (0) | 0 (0) | 0 (0) | – | 0 (0) | 0 (0) | 0 (0) | |
| AdV n (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | – | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| HBoV n (%) | 0 0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | – | 0 (0) | 0 (0) | |
| CoV 43 n (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | – | |
| Rhino+ADV+H1N1 n (%) | 1 (4) | 1 (4.2) | 1 (25) | |||||||
| RSV+AdV+Co43 n (%) | 1 (14.3) | 1 (25) | 1 (100) | |||||||
H1N1, influenza A(H1N1)pdm09; Rhino/EV, rhinovirus/enterovirus; RSV, respiratory syncytial virus; HMPV, human metapneumovirus; AdV, adenovirus; PIV, parainfluenza vírus; HBoV, human bocavirus; Flu, seasonal influenza vírus; Cor, coronavirus.
Of the 71 originally analyzed samples, 16 had incomplete data in the corresponding case report forms (22.5%). Therefore, most of the clinical variables and outcomes were evaluated in 55 cases.
Twenty-one patients (38.2%) presented with one comorbidity, and 28 (40%) with two or more comorbidities, such as chronic encephalopathy (25.6%), bronchiolitis (25.6%), cardiopathy (16.2%), hematological disease (16.2%), asthma (13.9%), malnutrition (13.9%), cancer (13.9%), other chronic pneumopathologies (9.3%), AIDS (4.6%), and obesity (2.3%).
Chest X-rays were abnormal in 96.4% of the patients, the most common being diffuse infiltrates (52.8%) and alveolar consolidations (43.4%).
Oseltamivir was administered to 70.9% of the cases, more than 48h after the onset of symptoms in most cases, while antibiotics were given in 85.5%, corticosteroids in 52.7%, and antifungal agents in 25.5% of the cases.
Organic dysfunction was present in 60% of the patients, and 90% of those had two or more affected systems.
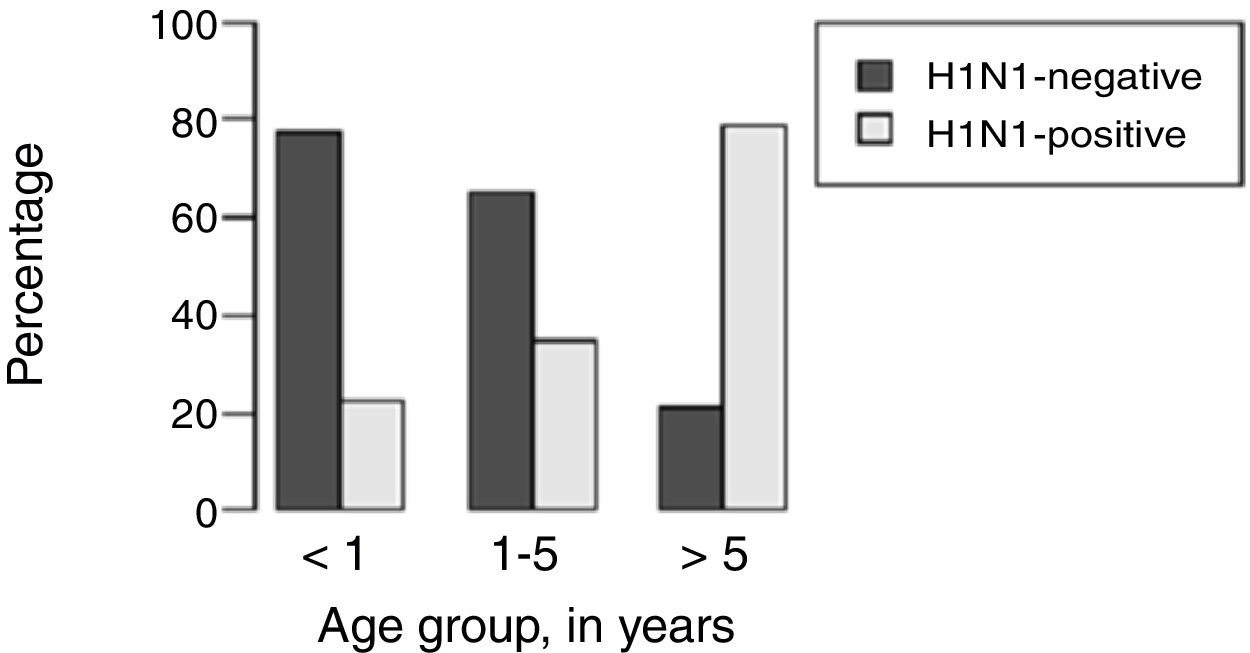
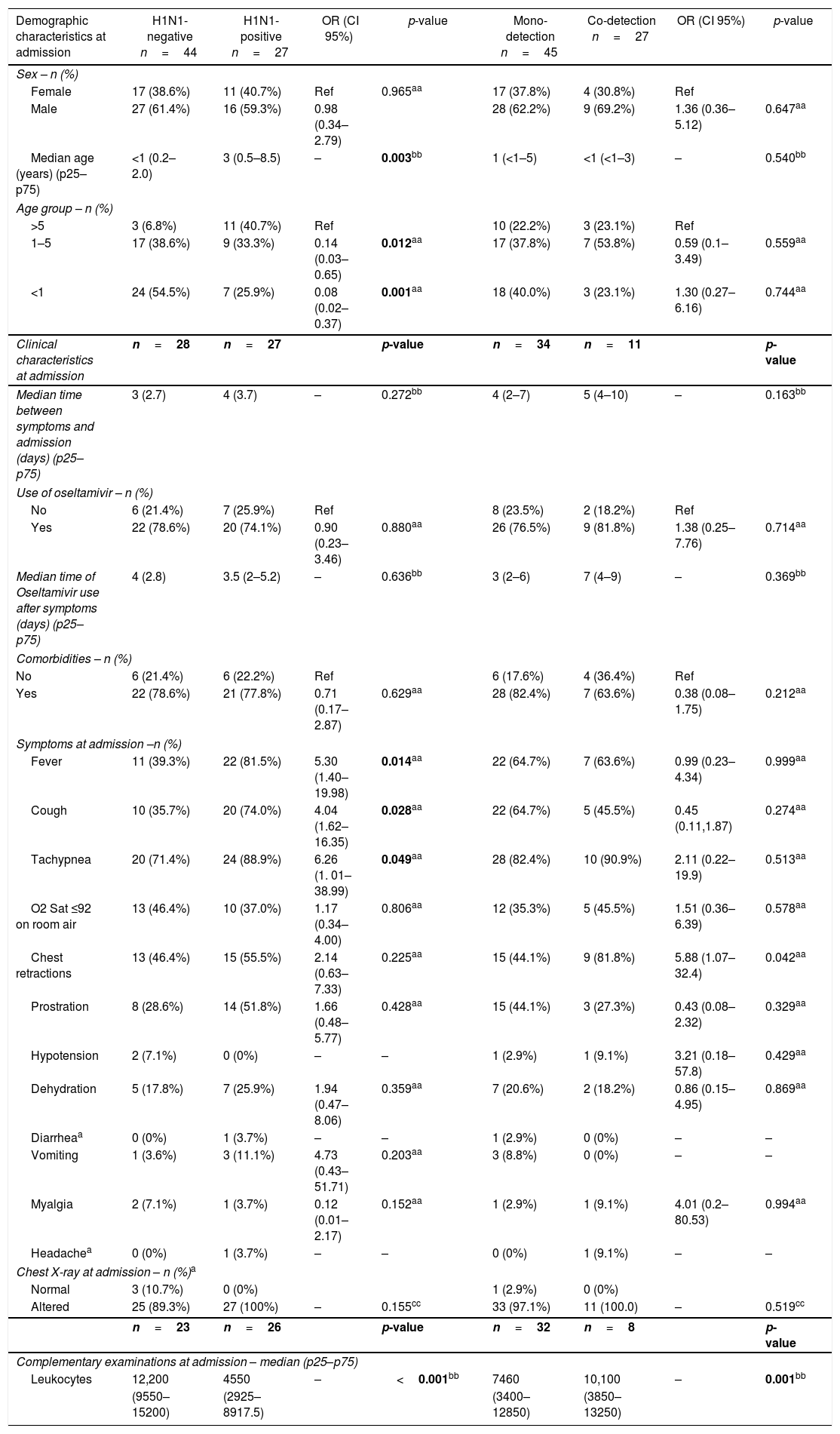
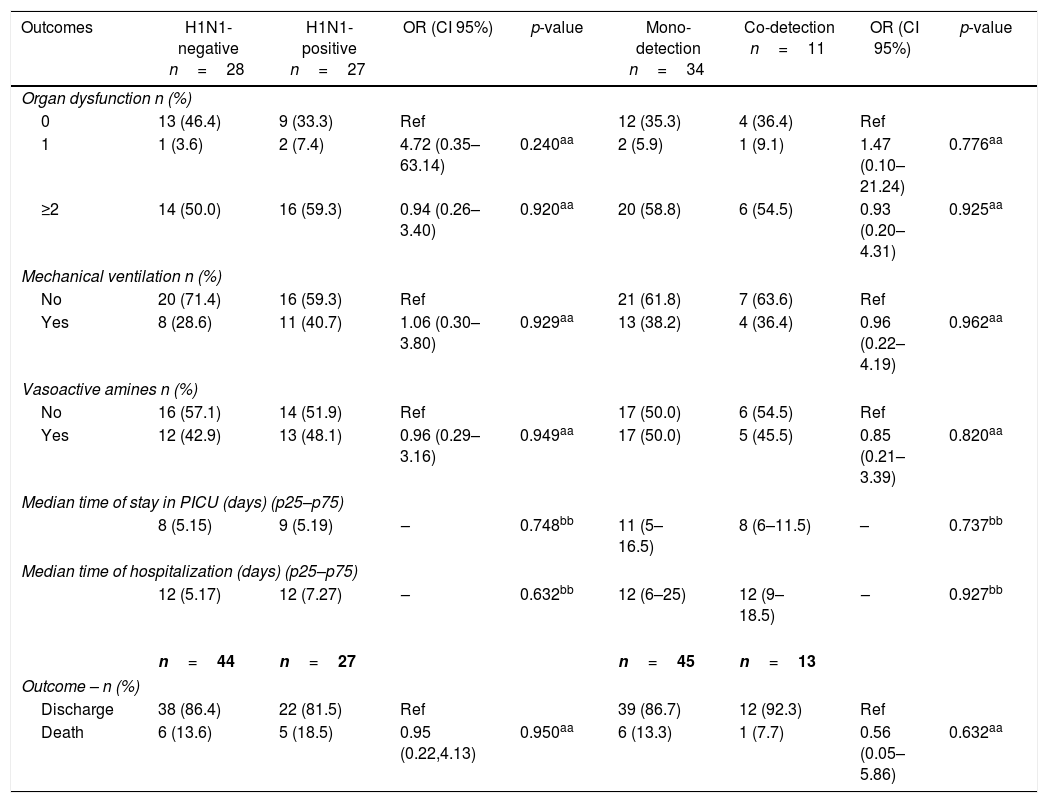
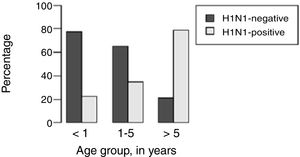
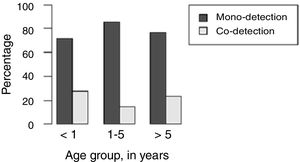
H1N1-positive compared to H1N1-negative patients had a higher median age (3 years vs. <1 year, p=0.003), but not codetection group compared to mono-detection group (Figs. 1 and 2 and Table 2). Therefore, all subsequent analyses were adjusted for age. H1N1-positive patients presented more frequently fever (OR=5.30, 95%CI=1.40–19.98, p=0.014), cough (OR=4.04, 95%CI=1.62–16.35, p=0.028), tachypnea (OR=6.26, 95%CI=1.01–38.99, p=0.049), and lower leukocyte count (median of 4550 vs. 12,200, p<0.001) (Table 4). There were no differences between the two groups in terms of organic dysfunction, use of mechanical ventilator and vasoactive amines, hospital and PICU length of stay, and death (Table 5).
Demographic and clinical characteristics at admission of children admitted to the PICU in Rio de Janeiro during the 2009 influenza A(H1N1) pandemic, according to H1N1-negative versus H1N1-positive detection and viral mono-detection versus viral co-detection in respiratory secretions.
| Demographic characteristics at admission | H1N1-negative n=44 | H1N1-positive n=27 | OR (CI 95%) | p-value | Mono-detection n=45 | Co-detection n=27 | OR (CI 95%) | p-value |
|---|---|---|---|---|---|---|---|---|
| Sex – n (%) | ||||||||
| Female | 17 (38.6%) | 11 (40.7%) | Ref | 0.965aa | 17 (37.8%) | 4 (30.8%) | Ref | |
| Male | 27 (61.4%) | 16 (59.3%) | 0.98 (0.34–2.79) | 28 (62.2%) | 9 (69.2%) | 1.36 (0.36–5.12) | 0.647aa | |
| Median age (years) (p25–p75) | <1 (0.2–2.0) | 3 (0.5–8.5) | – | 0.003bb | 1 (<1–5) | <1 (<1–3) | – | 0.540bb |
| Age group – n (%) | ||||||||
| >5 | 3 (6.8%) | 11 (40.7%) | Ref | 10 (22.2%) | 3 (23.1%) | Ref | ||
| 1–5 | 17 (38.6%) | 9 (33.3%) | 0.14 (0.03–0.65) | 0.012aa | 17 (37.8%) | 7 (53.8%) | 0.59 (0.1–3.49) | 0.559aa |
| <1 | 24 (54.5%) | 7 (25.9%) | 0.08 (0.02–0.37) | 0.001aa | 18 (40.0%) | 3 (23.1%) | 1.30 (0.27–6.16) | 0.744aa |
| Clinical characteristics at admission | n=28 | n=27 | p-value | n=34 | n=11 | p-value | ||
| Median time between symptoms and admission (days) (p25–p75) | 3 (2.7) | 4 (3.7) | – | 0.272bb | 4 (2–7) | 5 (4–10) | – | 0.163bb |
| Use of oseltamivir – n (%) | ||||||||
| No | 6 (21.4%) | 7 (25.9%) | Ref | 8 (23.5%) | 2 (18.2%) | Ref | ||
| Yes | 22 (78.6%) | 20 (74.1%) | 0.90 (0.23–3.46) | 0.880aa | 26 (76.5%) | 9 (81.8%) | 1.38 (0.25–7.76) | 0.714aa |
| Median time of Oseltamivir use after symptoms (days) (p25–p75) | 4 (2.8) | 3.5 (2–5.2) | – | 0.636bb | 3 (2–6) | 7 (4–9) | – | 0.369bb |
| Comorbidities – n (%) | ||||||||
| No | 6 (21.4%) | 6 (22.2%) | Ref | 6 (17.6%) | 4 (36.4%) | Ref | ||
| Yes | 22 (78.6%) | 21 (77.8%) | 0.71 (0.17–2.87) | 0.629aa | 28 (82.4%) | 7 (63.6%) | 0.38 (0.08–1.75) | 0.212aa |
| Symptoms at admission –n (%) | ||||||||
| Fever | 11 (39.3%) | 22 (81.5%) | 5.30 (1.40–19.98) | 0.014aa | 22 (64.7%) | 7 (63.6%) | 0.99 (0.23–4.34) | 0.999aa |
| Cough | 10 (35.7%) | 20 (74.0%) | 4.04 (1.62–16.35) | 0.028aa | 22 (64.7%) | 5 (45.5%) | 0.45 (0.11,1.87) | 0.274aa |
| Tachypnea | 20 (71.4%) | 24 (88.9%) | 6.26 (1. 01–38.99) | 0.049aa | 28 (82.4%) | 10 (90.9%) | 2.11 (0.22–19.9) | 0.513aa |
| O2 Sat ≤92 on room air | 13 (46.4%) | 10 (37.0%) | 1.17 (0.34–4.00) | 0.806aa | 12 (35.3%) | 5 (45.5%) | 1.51 (0.36–6.39) | 0.578aa |
| Chest retractions | 13 (46.4%) | 15 (55.5%) | 2.14 (0.63–7.33) | 0.225aa | 15 (44.1%) | 9 (81.8%) | 5.88 (1.07–32.4) | 0.042aa |
| Prostration | 8 (28.6%) | 14 (51.8%) | 1.66 (0.48–5.77) | 0.428aa | 15 (44.1%) | 3 (27.3%) | 0.43 (0.08–2.32) | 0.329aa |
| Hypotension | 2 (7.1%) | 0 (0%) | – | – | 1 (2.9%) | 1 (9.1%) | 3.21 (0.18–57.8) | 0.429aa |
| Dehydration | 5 (17.8%) | 7 (25.9%) | 1.94 (0.47–8.06) | 0.359aa | 7 (20.6%) | 2 (18.2%) | 0.86 (0.15–4.95) | 0.869aa |
| Diarrheaa | 0 (0%) | 1 (3.7%) | – | – | 1 (2.9%) | 0 (0%) | – | – |
| Vomiting | 1 (3.6%) | 3 (11.1%) | 4.73 (0.43–51.71) | 0.203aa | 3 (8.8%) | 0 (0%) | – | – |
| Myalgia | 2 (7.1%) | 1 (3.7%) | 0.12 (0.01–2.17) | 0.152aa | 1 (2.9%) | 1 (9.1%) | 4.01 (0.2–80.53) | 0.994aa |
| Headachea | 0 (0%) | 1 (3.7%) | – | – | 0 (0%) | 1 (9.1%) | – | – |
| Chest X-ray at admission – n (%)a | ||||||||
| Normal | 3 (10.7%) | 0 (0%) | 1 (2.9%) | 0 (0%) | ||||
| Altered | 25 (89.3%) | 27 (100%) | – | 0.155cc | 33 (97.1%) | 11 (100.0) | – | 0.519cc |
| n=23 | n=26 | p-value | n=32 | n=8 | p-value | |||
| Complementary examinations at admission – median (p25–p75) | ||||||||
| Leukocytes | 12,200 | 4550 | – | <0.001bb | 7460 | 10,100 | – | 0.001bb |
| (9550–15200) | (2925–8917.5) | (3400–12850) | (3850–13250) | |||||
The values of N and p-value with statistical significance are shown in bold.
OR, odds ratio; Ref, reference category.
Outcomes of severity in children admitted to the PICU in Rio de Janeiro during the 2009 influenza A(H1N1) pandemic, according to H1N1-negative versus H1N1-positive detection and viral mono-detection versus viral co-detection in respiratory secretions.
| Outcomes | H1N1-negative n=28 | H1N1-positive n=27 | OR (CI 95%) | p-value | Mono-detection n=34 | Co-detection n=11 | OR (CI 95%) | p-value |
|---|---|---|---|---|---|---|---|---|
| Organ dysfunction n (%) | ||||||||
| 0 | 13 (46.4) | 9 (33.3) | Ref | 12 (35.3) | 4 (36.4) | Ref | ||
| 1 | 1 (3.6) | 2 (7.4) | 4.72 (0.35–63.14) | 0.240aa | 2 (5.9) | 1 (9.1) | 1.47 (0.10–21.24) | 0.776aa |
| ≥2 | 14 (50.0) | 16 (59.3) | 0.94 (0.26–3.40) | 0.920aa | 20 (58.8) | 6 (54.5) | 0.93 (0.20–4.31) | 0.925aa |
| Mechanical ventilation n (%) | ||||||||
| No | 20 (71.4) | 16 (59.3) | Ref | 21 (61.8) | 7 (63.6) | Ref | ||
| Yes | 8 (28.6) | 11 (40.7) | 1.06 (0.30–3.80) | 0.929aa | 13 (38.2) | 4 (36.4) | 0.96 (0.22–4.19) | 0.962aa |
| Vasoactive amines n (%) | ||||||||
| No | 16 (57.1) | 14 (51.9) | Ref | 17 (50.0) | 6 (54.5) | Ref | ||
| Yes | 12 (42.9) | 13 (48.1) | 0.96 (0.29–3.16) | 0.949aa | 17 (50.0) | 5 (45.5) | 0.85 (0.21–3.39) | 0.820aa |
| Median time of stay in PICU (days) (p25–p75) | ||||||||
| 8 (5.15) | 9 (5.19) | – | 0.748bb | 11 (5–16.5) | 8 (6–11.5) | – | 0.737bb | |
| Median time of hospitalization (days) (p25–p75) | ||||||||
| 12 (5.17) | 12 (7.27) | – | 0.632bb | 12 (6–25) | 12 (9–18.5) | – | 0.927bb | |
| n=44 | n=27 | n=45 | n=13 | |||||
| Outcome – n (%) | ||||||||
| Discharge | 38 (86.4) | 22 (81.5) | Ref | 39 (86.7) | 12 (92.3) | Ref | ||
| Death | 6 (13.6) | 5 (18.5) | 0.95 (0.22,4.13) | 0.950aa | 6 (13.3) | 1 (7.7) | 0.56 (0.05–5.86) | 0.632aa |
OR, odds ratio; CI, confidence interval; Ref, reference category.
Comparing mono-detection and codetection groups, no differences in relation to variables at admission or outcomes were found (Tables 4 and 5).
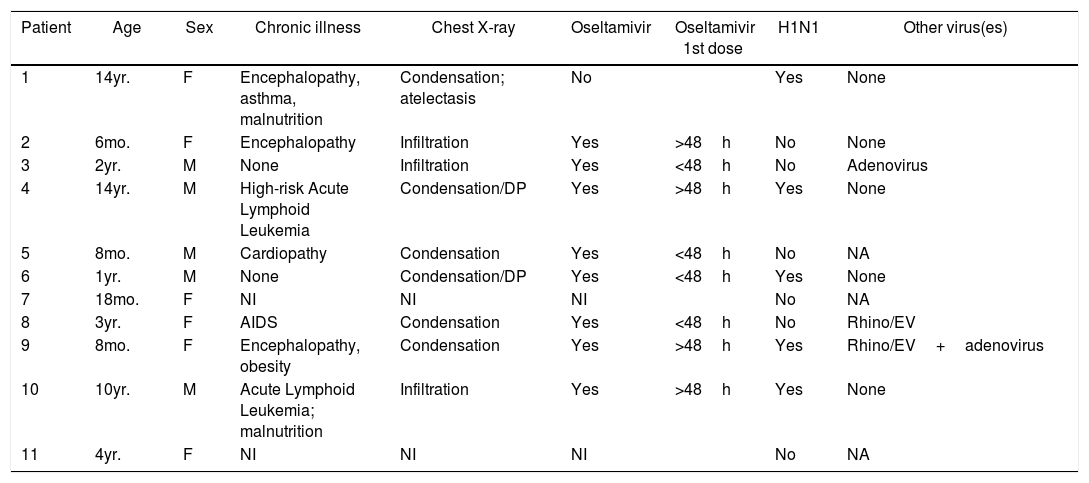
The fatality rate was 15.5%. Of the 11 patients who died, clinical data forms were only available for nine. Seven had comorbidities, and all nine had abnormalities in chest X-rays. Eight patients were given oseltamivir, but only four within 48h of the onset of symptoms. H1N1 virus was isolated from five of these 11 patients, in four cases as a single virus and in one case associated to RV/EV and adenovirus. In the remaining six samples, adenovirus was detected in one, RV/EV in another and no viruses in a third. (Table 6).
Characteristics of patients with an outcome of death, among the 71 children admitted to the PICU in Rio de Janeiro during the 2009 influenza A(H1N1) pandemic.
| Patient | Age | Sex | Chronic illness | Chest X-ray | Oseltamivir | Oseltamivir 1st dose | H1N1 | Other virus(es) |
|---|---|---|---|---|---|---|---|---|
| 1 | 14yr. | F | Encephalopathy, asthma, malnutrition | Condensation; atelectasis | No | Yes | None | |
| 2 | 6mo. | F | Encephalopathy | Infiltration | Yes | >48h | No | None |
| 3 | 2yr. | M | None | Infiltration | Yes | <48h | No | Adenovirus |
| 4 | 14yr. | M | High-risk Acute Lymphoid Leukemia | Condensation/DP | Yes | >48h | Yes | None |
| 5 | 8mo. | M | Cardiopathy | Condensation | Yes | <48h | No | NA |
| 6 | 1yr. | M | None | Condensation/DP | Yes | <48h | Yes | None |
| 7 | 18mo. | F | NI | NI | NI | No | NA | |
| 8 | 3yr. | F | AIDS | Condensation | Yes | <48h | No | Rhino/EV |
| 9 | 8mo. | F | Encephalopathy, obesity | Condensation | Yes | >48h | Yes | Rhino/EV+adenovirus |
| 10 | 10yr. | M | Acute Lymphoid Leukemia; malnutrition | Infiltration | Yes | >48h | Yes | None |
| 11 | 4yr. | F | NI | NI | NI | No | NA |
NI, no information; NA, not applicable (only one method used).
In the present study, 38% of all samples were positive for H1N1 virus, while 92.1% were positive for some virus. Viral codetection occurred in 20% of the H1N1-positive samples and in 22.5% of all positive samples for some virus. H1N1-positive patients were older, had higher frequency of fever and cough, and lower leukometry compared to H1N1-negative patients. We found no association between viral codetection and clinical variables or outcomes.
Data on the profile and frequency of viral detection and codetection with H1N1 virus are highly variable among the studies.4,7,9,12,15
The 2009 pandemic presented unique opportunity to study the H1N1 virus, other etiological agents implicated in SARI, and viral codetection. There are reports of positive nonspecific viral detection ranging from 60 to 90% during the 2009 pandemic.8,9,12,15 The high frequency of virus positivity observed in our study in the retested samples (92.1%) can probably be explained by the association of methods used to test most samples and the high sensitivity and specificity of the laboratory methods employed16–18 since three or more methods were employed in 81% of the retested samples and the RespiFinder and/or Fast-track methods were used in 97% of the retested samples.
The frequency of H1N1 detection in our study (38%) was consistent with literature data of hospitalized patients during the epidemic in other cities in Brazil, which ranged from 18.2% in São Paulo (children and adults)8 to 31.1% in Porto Alegre (only children).10 In other countries, frequencies of H1N1 detection in hospitalized patients varied from 16.5% in Sweden (only children)12 to 47% in the US (children and adults).4
The frequency of viral codetection in previous reports also varies widely, but is higher in young children. In USA, two studies of hospitalized adults and children found a viral codetection rate of 7.3%4 and 13%.7 In hospitalized children, the frequency of coinfection was 14.6% in Sweden,12 41% of preschool-age patients in Brazil,9 and 61.8% in Barcelona.15
In our study, viral codetection was more frequent in H1N1-negative patients, who were younger than H1N1-positive patients. The most frequent codetection was a combination of RV-EV and RSV. Similar results were found in Barcelona, where the main codetections were RV-EV/RSV-B and RSV-A/RSV-B.15
The high frequency of comorbidities in the population of critically ill children of our study, primarily chronic encephalopathy and a previous history of bronchiolitis, was similar to that reported in hospitalized patients in the US (73%)19 and in children admitted to the PICU in Germany (75%),20 where neurodevelopmental diseases were the most prevalent.
The main symptoms found in our study group were tachypnea, followed by fever and cough. The H1N1-positive subgroup presented a higher frequency of fever, cough and tachypnea and lower leukocyte count when compared to the H1N1-negative subgroup. Cough is a characteristic of H1N1 infection,21–23 as well as leukopenia.9 In Taiwan, leukopenia was found in 64.2% of cases and was more frequent in H1N1-positive.22
During the 2009 pandemic, H1N1 was more frequent in young adults and in children younger than five years of age.6,24–26 In addition, the incidence of SARI for H1N1 was higher in children younger than two years24 and this group was included as a priority in the first vaccination campaigns in Brazil.13 This phenomenon can be explained by the high susceptibility of children to H1N1, a virus with a high potential for transmission and completely new to the pediatric population, who had no antibodies with cross-reactivity.27,28 In our study, however, H1N1-positive children were older than the H1N1-negative children. Although this may have been only an effect of the small sample size, other studies have found similar results.6,21,29 The main hypothesis raised by some of these authors is that the presence of previous acquired non-protective antibodies in older children and young adults might lead to a cross-reaction with the H1N1 virus, which could lead to immunecomplex formation and severe lung disease.29 Smaller children would have a less robust immune response to H1N1 virus, preventing progression to immune-mediated lung disease.21 Another possible explanation would be an earlier and more aggressive treatment in younger children, because they are a risk group.21
In our study, chest X-ray imaging showed diffuse infiltrates in 52.8% of the cases and alveolar consolidations in 43.4%, which is consistent with previously published studies.4
We observed a high case fatality rate of 15.5%, possibly related to the severity of illness in our study population, which is comparable to literature data. A fatality rate of 18.6% was found in a German study with a similar population of PICU-admitted children during the pandemic period.20 In Porto Alegre, Brazil,10 a fatality rate of 9.1% was described among younger hospitalized children.
The association between viral codetection and severity of illness in children is controversial in the literature. Some studies call attention to specific codetections associated with worse prognosis.29,30 We compared the H1N1-positive and negative groups, as well as the nonspecific viral codetection and mono-dectection groups in relation to severity outcomes. We found no association between these outcomes and the presence of H1N1 or nonspecific viral codetection in respiratory secretions, but our small sample size may have influenced the strength of causal association. In other Brazilian studies carried out during the pandemic, the authors found an association of viral codetection with hospitalization9and mechanical ventilation.10 In Ohio, patients coinfected with rhinovirus tended to present a less severe clinical picture than those coinfected with other viruses.7
Data from epidemiological reports in Brazil31–33 and studies in other countries3,34,35 showed that, in the post-pandemic period, H1N1 virus continues to circulate and cause SARI and deaths. The influenza virus continues to mutate and frequently re-appears in variant forms. In March 2013, the influenza A(H7N9) virus circulated mainly in China causing serious illness.36 In November 2015, there was one human case of influenza A(H1N2) in Brazil.33 The threat of new pandemics is a reality and continuing surveillance is essential.
This study has some limitations. The use of retrospective data restricted the amount of information available. The incomplete data allied to the small sample size made comparisons difficult between subgroups. The non-significant associations between the analyzed subgroups and the studied variables and outcomes may have occurred due to lack of statistical power.
In addition, the viral isolation methods were performed according to availability of reagents and equipment, resulting in the utilization of four different methods. However, most samples were tested with the RespiFinder and/or Fast-track methods, optimizing virus isolation. Finally, there was a great interval between the first test for H1N1 and the retest for other virus. Nevertheless, the samples were stored appropriately and were retested only after an assessment for viability.
ConclusionsOther etiologic agents besides influenza A(H1N1)pmd09 virus can be responsible for SARI in children admitted to the PICU. The association between viral codetection and disease severity is still controversial. This study represents a contribution to the understanding of viral respiratory infections in the poorly studied population of children admitted to the PICU. More epidemiological studies and improvements in viral detection techniques are needed to define the true role of viral coinfections in respiratory diseases.
Ethical approvalThis study was approved by the Committee of Ethics in Research of the Institute for Research & Education (IDOR), Rio de Janeiro, Brazil, on September 23, 2014 (CAEE: 34179214.3.0000.5249). The consent term was waived since this was a retrospective data extraction from patients records.
FundingFundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Grant Number E-26/110.753/2010.
Conflicts of interestThe authors declare no conflicts of interest.