One of the hallmarks of COVID-19 is overwhelming inflammation, which plays a very important role in the pathogenesis of COVID-19. Thus, identification of inflammatory factors that interact with the SARS-CoV-2 can be very important to control and diagnose the severity of COVID-19. The aim of this study was to investigate the expression patterns of inflammation-related non-coding RNAs (ncRNAs) including MALAT-1, NEAT-1, THRIL, and miR-155-5p from the acute phase to the recovery phase of COVID-19.
MethodsTotal RNA was extracted from Peripheral Blood Mononuclear Cell (PBMC) samples of 20 patients with acute COVID-19 infection and 20 healthy individuals and the expression levels of MALAT-1, NEAT-1, THRIL, and miR-155-5p were evaluated by real-time PCR assay. Besides, in order to monitor the expression pattern of selected ncRNAs from the acute phase to the recovery phase of COVID-19 disease, the levels of ncRNAs were re-measured 6‒7 weeks after the acute phase.
ResultThe mean expression levels of MALAT-1, THRIL, and miR-155-5p were significantly increased in the acute phase of COVID-19 compared with a healthy control group. In addition, the expression levels of MALAT-1 and THRIL in the post-acute phase of COVID-19 were significantly lower than in the acute phase of COVID-19. According to the ROC curve analysis, these ncRNAs could be considered useful biomarkers for COVID-19 diagnosis and for discriminating between acute and post-acute phase of COVID-19.
DiscussionInflammation-related ncRNAs (MALAT-1, THRIL, and miR-150-5p) can act as hopeful biomarkers for the monitoring and diagnosis of COVID-19 disease.
The Coronavirus Disease Pandemic of 2019 (COVID-19) is caused by a novel coronavirus known as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). At the moment, 239 million individuals have been infected with SARS-CoV-2, and much more than 4.8 million have died as a result of this infection.1 In addition, the epidemic has triggered worldwide social and economic instability. SARS-CoV-2 is an enveloped virus with a positive non-segmented single-stranded RNA genome which belongs to Nidovirales order, Coronaviridae family, Betacoronavirus genus, and lineage B.2 Because of the worldwide severity of the pathogen, the increased infection rate of SARS-CoV-2, and the limitation of effective treatment approaches, more research is needed to properly control their spread and to offer treatment alternatives.
Over than 90% of the human DNA sequence is constantly transcribed, but only 2% of it produces proteins. The vast majority of transcripts are classified as non-coding RNAs (ncRNAs). According to their sequence length, ncRNAs are classified into long non-coding RNA (lncRNA) with size larger than 200 nucleotides (nt) and small non-coding RNA (sncRNA) with length less than 200 nt, such as microRNAs, which are both important epigenetic and sub-cellular regulatory elements that can be involved in complex cellular biological processes.3 Reportedly, lncRNAs may play essential regulatory functions in the interaction between virus and host, including regulation of host antiviral responses, direct and indirect roles in viral and host gene transcription, as well as regulation of the stability and translation of mRNAs.4 Also, it has been found that viral proteins can influence the expression level of cellular lncRNAs and microRNAs (miRNAs). As a result, changes in the expression level of these factors, directly and/or indirectly, can affect viral infection through regulating host innate immune responses, such as inflammation, and by regulating expression of both cellular and viral genes.5-7
MicroRNA-155-5p (miR-155-5p) has been characterized as an ancient immune cell regulator. MiR-155-5p is remarkable in the immune system because it can change the transcription of activated myeloid and lymphoid cells, regulating a wide range of biological processes from inflammation to immune response.8 Numerous research in recent years have demonstrated that miR-155-5p is evolutionarily conserved. Its expression is continuously increased in diverse cellular systems during viral infections in both animal and human models.9,10 Similarly, in animal models of Acute Respiratory Distress Syndrome (ARDS), increased miR-155-5p is associated with respiratory infections, illness severity, and greater mortality.11,12 Overall, evidence strongly suggests that miR-155 has a critical role as regulator of inflammation13 and during most viral infections, since the expression level of miR-155 is upregulated and regulates antiviral immune responses.14
Nuclear Factor-kappa B (NF-κB) is a dimeric transcription factor involved in inflammation and has an important role in pathogenesis of several inflammatory disease such as Chronic Obstructive Pulmonary Disease (COPD) and COVID-19.15 Numerous ncRNAs such as miR-155-5p, MALAT-1, NEAT-1 and THRIL are involved in regulating the NF-κB signaling pathway.16 It has been reported that lncRNA MALAT-1 can control cytokine secretion in macrophages under inflammatory circumstances and promote inflammatory activity by interacting with the NF-κB signaling pathway.17,18 NEAT-1 is another lncRNA that has been shown to play a role in NF-κB signaling pathway and NEAT-1 inhibition prevented the activation of the NF-κB pathway.19 and induced expression of inflammatory-related cytokines such as IL-8 and IL-6.20 Reportedly, NEAT-1 probably can help the inflammation-regulating ncRNA-mRNA network, and some factors linked with this network may be able to regulate inflammation by interacting with essential inflammatory mediators such as IL-6, TNF and muscarinic acetylcholine receptors.21-23 THRIL is a newly described lncRNA that has been confirmed to interact with hnRNPL (Heterogeneous Nuclear Ribonucleoprotein L) and then controlling the expression of TNF-α with an important role in regulation of inflammation and immune response.24,25 THRIL induce the upregulation of NRP1 expression and further induce the modulation of the NF-κB signaling pathway.26 As a result, the interaction between lncRNAs and targets (e.g., miRNAs, cellular factors and viral genes) has sparked researchers' interests to investigate the potential biomarkers and/or therapeutic targets. Currently, our understanding of SARS-CoV-2 processes is limited, and there are no particular biomarkers associated with SARS-CoV-2 diagnosis or therapy. Since selected cellular ncRNAs (miR-155-5p,27-31 MALAT-1,32-35 NEAT-120,36,37 and THRIL38,39) may play critical roles in immune response regulation and inflammation, we evaluated the expression pattern of lncRNAs (MALAT-1, NEAT-1 and THRIL) and miR-155-5p in Peripheral Blood Mononuclear Cells (PBMC) of SARS-CoV-2 infected individuals in both acute and post-acute stages and compared to healthy individuals.
Patients and methodsPatients’ selectionFrom June 2021 to July 2021, 20 patients with COVID-19 infection were recruited from the West Health Center in Tehran (related to Iran University of Medical Sciences [IUMS]) and enrolled in this cross-sectional survey. A peripheral blood sample of 6 mL was collected from these patients during the acute phase and again in the post-acute phase, and from 20 healthy controls.
It should be noted that the studied participants did not have co-infections with Human Immunodeficiency Virus (HIV), Human Cytomegalovirus (HCMV), Hepatitis B Virus (HBV), and Hepatitis C Virus (HCV), and Mycobacterium tuberculosis. Furthermore, none of the subjects had underlying medical conditions.
Ethical issuesThis study was approved by the ethics committee of IUMS (ethical code: IR. IUMS. REC.1400.381), and all of the participants filled written informed consent for blood specimen collection.
Preparation of peripheral blood mononuclear cells (PBMCs)Collected peripheral blood from each subject was transferred into a tube containing Ethylenediaminetetraacetic Acid (EDTA) as anticoagulant and then separated by centrifugation. PBMCs were isolated based on the ficoll hypaque density gradient centrifugation (Lympholyte-H, Cedarlane, Hornby, Canada) technique according to the manufacturer's instructions, and then the pellet of PBMCs was washed three times with phosphate-buffered saline (pH: 7.3±0.1), and finally re-suspended with 350 µL of RNA maintenance solution (RNA-Later [Ambion, Inc., Austin, TX]), and kept at -80°C until extraction of the total RNA.
Total RNA isolation and complementary DNA (cDNA) synthesisTotal RNA was extracted from PBMC samples according to the manufacturer's protocols with minor modifications. Briefly, after PBMC lysis with 1 mL QIAzol solution, 250 μL of chloroform was added to the lysate, shaken vigorously for one minute, and after 5‒10 minutes of incubation at room temperature, centrifuged at 12,000 × g for 15 minutes at 4°C.The supernatant was aspirated and approximatel 800 μl isopropanol was added and placed in the freezer overnight. The samples were centrifuged at 12,000 × g for 45 minutes at 4°C. One mL ethanol (100%) was added to the RNA pellet and the microtubes went up and down several times and centrifuged at 12,000 × g for 15 minutes at 4°C. The pellet of RNA was air-dried and dissolved with RNase/DNase free distilled water. The integrity and purity of the isolated RNA was evaluated using a Nano-Drop spectrophotometer (Thermo Scientific, Wilmington, MA) instrument, and then kept at -80°C until the test.
To determine the expression pattern of lncRNAs (MALAT-1,40 NEAT-141 and THRIL,42), as well as GAPDH and β-actin (as normalization controls for relative quantification),42,43 cDNA was synthesized using 350 ng of the total RNA as previously described in detail.44
Expression analysis of genes using real-time PCRThe expression patterns of lncRNAs (MALAT-1,40 NEAT-141 and THRIL42), and also GAPDH (the expression level of this housekeeping gene was considered as reference gene)45 were determined by real-time Polymerase Chain Reaction (PCR) using a Rotorgene Q thermal cycler (Qiagen, Hilden, Germany) instrument. The assays were done on 20 μL reaction mixture including: 10 pmol of each primer (MALAT-1, NEAT-1, THRIL, GAPDH and β-actin), 8 μL nuclease free distilled water, 10 μL 2 × SYBR® Premix Ex Taq (Tli Plus) Master Mix (TaKaRa Bio Inc. Shiga, Japan) (Table 1) and one μL of cDNA as template.
Primers used in this study for determining of expression profile of long non-coding RNAs (lncRNAs).
MALAT-1, Metastasis Associated Lung Adenocarcinoma Transcript 1; NEAT-1, Nuclear Paraspeckle Assembly Transcript 1; THRIL, TNF and HNRNPL Related Immunoregulatory Long non-coding RNA; GAPDH, Glyceraldehyde 3-Phosphate Dehydrogenase.
The thermocycling conditions for real-time PCR were defined as follows: initial denaturing at 95°C for 15 minutes, and 40 cycles, including 15 seconds at 95°C, 30 seconds at 60°C, and 20 seconds at 72°C. The 2−ΔΔCT method was used for calculation of the relative expression values. All the specimens were tested in duplicate reactions.
miRNA-155-5p expression analysisTotal RNA was extracted (as described in the previous section) from PBMC samples. Complementary DNA (cDNA) was synthesized using 5 μg of the total RNA as previously described in detail.46 In the current study the expression of miR-155-5p was evaluated in PBMC specimens of SARA-CoV-2 infected patients in the two stages of disease (acute and post-acute phase) and of healthy controls based on available information.
The real time PCR assay was carried out in final 20 μL volume, including 0.5 μL of specific forward primer, 0.5 μL of universal reverse primer, 10 μL of SYBR Green PCR Master Mix (TaKaRa, Kusatsu, Japan), 8 μL of nuclease-free water, and one μL of cDNA as template. The thermal profile of this assay (three steps with melt) was set at 95°C for two minutes as hold time, followed by 40 cycles of denaturation at 95°C for 15 seconds, annealing at 60°C for 20 seconds, and extension at 72°C for 25 seconds, and also melting curve analysis was determined at temperatures ranging from 55 to 99°C. This assay was performed using the Rotorgene Q thermal cycler (Qiagen, Hilden, Germany) instrument. The expression levels of miR-155-5p was normalized to Snord47 and 68 as reference RNA and the fold change was calculated by the Livak method.47 It should be noted that all reactions were done in triplicate.
Statistical analysisData were analyzed using SPSS version 16 (SPSS Inc., Chicago, IL, USA) and Prism 6.0 software (GraphPad, San Diego, CA, USA). Clinical and demographics characteristics were presented as n (%) for categorical variables and mean ± Standard Deviations (SD) for age, which were analyzed by Fisher's exact test and Student t-Test, respectively. The Mann-Whitney U-test or the independent-samples t-test was used to compare the mean expression levels of ncRNAs (MALAT-1, NEAT-1, THRIL and miR-155-5p) between the COVID-19 groups with the healthy control group. The statistical difference of the mean level of ncRNAs between acute and post-acute phases of COVID-19 disease was compared using the paired sample t-test. The Receiver Operating Characteristic (ROC) curve analysis was performed to evaluate the diagnostic value of ncRNA expression level in discriminating between study groups. The Spearman rank correlation was used to compare the association of variables. The Benjamini and Hochberg procedure was used to control for the false discovery rate. All statistical evaluations were two-tailed, and p-values less than 0.05 were considered significant.
ResultsCharacteristics of participantsFifty SARS-CoV-2 infected patients (in both acute and post-acute phases of the disease) and 50 healthy individuals were enrolled in this cross-sectional study. These two studied groups were matched for sex and age. The mean age of studied patients with SARS CoV-2 infection was 36.1±10.9 (ranging between 22‒67 years) and for healthy individuals was 36.2±12.1 (ranging between 23‒67 years). The demographic parameters of the studied participants and clinical manifestations of patients with SARS-CoV-2 infection are summarized in Table 2.
The demographic parameters of the studied participants and clinical manifestations of patients with SARS-CoV-2 infection.
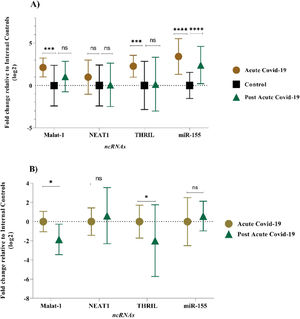
The expression level of MALAT-1, NEAT-1, THRIL, and miR-155-5p were examined in PBMC samples during acute and post-acute COVID-19 disease and healthy control subjects. In acute COVID-19 group, expression profiles of MALAT-1, THRIL and miR-155-5P were found significantly deregulated (p-value of < 0.05) when compared with healthy controls. In comparison to the control group, the acute COVID-19 group showed higher expression levels: 3.42-fold for the miR-155-5p, 2.27-fold for THRIL, and 2.12-fold for MALAT-1, respectively. Also, there was no significant difference in the mean expression level of NEAT-1 between acute COVID-19 group and control group (Fig. 1A), (p-value > 0.05). The expression pattern of lncRNAs (MALAT-1, NEAT-1 and THRIL) in post-acute COVID-19 group was similar to the healthy control group, as shown in Figure 1A. However, the mean expression level of miR-155-5p in post-acute COVID-19 was higher than that of control subjects (2.49-fold change, p-value < 0.0001).
In order to identify a PBMC biomarker that could be applicable to distinguish the prognosis of COVID-19 disease, the PBMC level of the selected lncRNAs (MALAT-1, NEAT-1 and THRIL) and miR-155-5p were re-measured 6‒5 weeks after the acute phase of COVID-19. Mean expression level of MALAT-1 and THRIL were significantly down-regulated (-1.8-fold, p-value = 0.037 and -1.98-fold, p-value = 0.022, respectively) in post-acute phase of COVID-19 compared to acute phase of COVID-19 disease (Fig. 1B). However, there was no significant difference in the expression pattern of miR-155-5p, and THRIL between acute COVID-19 group and post-acute COVID-19 group (p-value > 0.05).
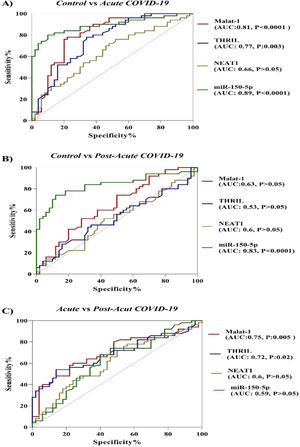
In the current study, the potential of ncRNAs (miR-155-5p, MALAT-1, NEAT-1 and THRIL) in discriminating between COVID-19 groups and healthy controls and between acute phase of COVID-19 and post-acute phase of COVID-19 was evaluated by the ROC curve analysis. According to results which are illustrated in Figure 2, miR-155-5p (AUC = 0.89, p-value < 0.0001), MALAT-1 (AUC = 0.81, p-value < 0.0001) and THRIL (AUC = 0.77, p-value = 0.003) are effective in distinguishing acute phase of COVID-19 from healthy controls. In the case of post-acute COVID-19 phase compared with healthy controls, the AUC value for miR-155-5p was 0.83 (p-value < 0.0001). Especially, the miR-155-5p showed an excellent AUC value. Lastly, PBMC THRIL and MALAT-1 were able to distinguish acute phase of COVID-19 from post-acute phase of COVID-19 disease with AUC value of 0.75 (p-value = 0.005) and 0.72 (p-value = 0.021), respectively (Fig. 2).
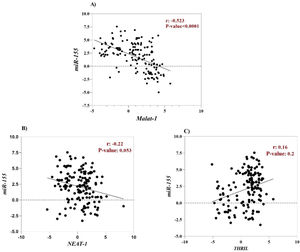
The correlation between clinical characteristics and relative expressions of ncRNAsHsa-miR-155-5p is one of the cellular miRNAs that maybe play a critical role in regulating inflammation and antiviral cellular defense in SARS-CoV-2 infection.48,49 Furthermore, it has been documented that some cellular lncRNAs, such as MALAT-1, cause modulating miR-155-5p expression.49 For this reason, to determine the correlation between the expression level of lncRNAs (MALAT-1, NEAT-1 and THRIL) and miR-155-5p, Spearman correlation analysis was performed. According to the result, a negative correlation was found between MALAT-1 level and level of miR-155-5p (r = -0.52, p-value < 0.0001), (Fig. 3A). However, there was no significant correlation between expression level of NEAT-1 and THRIL with miR-155-5p expression level (Fig. 2 B and C).
To investigate the relationship among expression level of ncRNAs with demographic-clinical characteristics and by SARS-CoV-2 (RdRP and N) genes in the COVID-19 patients during acute COVID-19 disease, Spearman correlation coefficient was carried out (Table 3). According to the result, there was a significant negative correlation between delta Ct of miR-155-5p and delta Ct of RdRp (r = -0.7, p-value < 0.001) and N genes (r = -0.61, p-value < 0.01) of SARS-Cov-2. Besides, a significant positive correlation was found among delta Ct of THRIL and NEAT-1 with dry cough (r = 0.57, and r = 0.46, respectively) and with sputum cough (r = 0.42 and r = 0.58, respectively). As well, a significant positive correlation was found between MALAT-1 and fever (r = 0.61, p-value < 0.001), skeletal pain (r = 0.55, p-value < 0.01) More information is provided in Table 3.
Spearman's correlation coefficient between the expression level of ncRNAs with expression level of SARS-CoV-2 genes (N and RdRp genes), demographic and clinical characteristics.
| MALAT-1 | NEAT-1 | THRIL | miR-155-5p | |
|---|---|---|---|---|
| RdRp | 0.43ns | 0.28ns | 0.4ns | -0.7c |
| N gene | 0.35ns | 0.24ns | 0.33ns | -0.61b |
| Sex Age | 0.09ns | 0.17ns | 0.07ns | -0.05ns |
| Bleeding stomach | 0.11ns | 0.28ns | 0.28ns | -0.08ns |
| Gastrointestinal symptoms | 0.37ns | 0.02ns | 0.16ns | 0.08ns |
| Decreased taste | -0.33ns | 0.45a | 0.04ns | 0.02ns |
| Decreased smell | -0.05ns | 0.07ns | 0.14ns | -0.18ns |
| Cape of nose | 0.12ns | 0.3ns | 0.41a | -0.07ns |
| Runny nose | 0.1ns | 0.24ns | 0.36ns | -0.19ns |
| Shortness of breath | -0.1ns | 0.38ns | 0.38ns | 0.36ns |
| Chest pain | 0.07ns | 0.43ns | 0.46a | 0.3ns |
| Sputum cough | -0.31ns | 0.58b | 0.42a | 0.41a |
| Dry cough | -0.2ns | 0.46a | 0.57b | -0.31ns |
| Skeletal pain | 0.55b | 0.13ns | 0.2ns | 0.001ns |
| Chills | 0.28ns | 0.17ns | 0.39ns | 0.13ns |
| Headache | -0.18ns | -0.2ns | 0.13ns | 0.33ns |
| Confusion | 0.07ns | 0.36ns | 0.17ns | 0.13ns |
| Fever | 0.61c | 0.1ns | -0.004ns | 0.2ns |
Increasing evidence indicated that ncRNAs play a critical role in inflammation-related disorders by regulation of the diverse biological processes, such as the activation of inflammatory pathway signaling.50,51 Reportedly, ncRNAs cross-talking with immune cells can also regulate inflammation and immunological response. Further, due to growing understanding of the interactions between SARS-CoV-2 with host ncRNAs,52,53 one could suggest that SARS-CoV-2 can alter the immunological pathway by deregulation of ncRNAs expression. Result of this research point out that the expression level of MALAT-1, NEAT-1, THRIL, and miR-155-5p, which are associated with inflammatory response, were significantly different between COVID-19 patients and healthy control subjects, as well as between acute and post-acute phase of COVID-19 disease. Recently, it was shown that expression level of MALAT-1 was significantly upregulated in SARS-CoV-2-infected bronchial epithelial cells.54 In another study, Huang et al.55 reported that expressions of NEAT-1 and MALAT-1 were significantly increased in severe COVID-19 patients compared to mild COVID-19 patients and they suggested that NEAT-1 and MALAT-1 promote cellular damage and stress.55 In addition, an in vivo study reported that silencing MALAT-1 inhibited neutrophil chemotaxis by interleukin-8 and suppresses pulmonary epithelial cells apoptosis.56 All of these findings suggest that MALAT-1 is increased in lung cells of COVID-19 patients, promoting immune cell taxi and subsequent harmful inflammation. MALAT-1 expression is also linked to macrophage activation and maturation into the M1 subtype, which is important in numerous pathological events, including inflammation.57 Besides, MALAT-1 can promote the expression of Maf and IL-10 in T-helper (Th) cells and eventually suppresses immunity against infection.58 Recently, Rodrigues et al.59 investigated the expression of miR-3142, MALAT-1, and NEAT-1 in nasopharyngeal swab and saliva specimens of COVID-19 patients. They observed that expression levels of the NEAT-1 and MALAT-1 in SARS-CoV-2 positive samples were higher than those of healthy controls. Further, they suggested that salivary NEAT-1 could act as a potential biomarker for distinguishing between healthy subjects from COVID-19 patients (AUC = 0.80).59 Similarly, our data reveal that the level of MALAT-1 was significantly overexpressed in the acute phase of COVID-19 disease compared to healthy subjects. However, the expression level of MALAT-1 was significantly decreased from the acute phase to the post-acute phase of COVID-19 disease.
NEAT-1, a pro-inflammatory lncRNA which is comparable genomically to MALAT-1, was found to increase inflammation through enhanced inflammasome assembly and processing.60 NEAT-1 promote inflammation by induction of inflammatory cytokines such as Interleukin-6 (IL-6). In response to SARS-CoV-2 infection, IL-6 is one of the key immune components.59 In nine cell types (M1 and M2 type macrophages, monocytes, CD4+ T cells, and CD8+ memory T cells) identified from severe COVID-19 patient Bronchoalveolar Lavage (BAL) samples, there was overexpression of NEAT-1.61 According to our findings, no significant difference was observed in PBMC level of NEAT-1 between the acute COVID-19 group and the control group, and also the mean expression level of NEAT-1 during the acute phase of COVID-19 was statistically similar to the post-acute phase of the disease. These results suggest that the immunological effect of NEAT-1 may be specific to the lung, i.e., where the infection and inflammation initiate. Besides, different results from previous studies were possible because of differences in the type of samples.
The lncRNA THRIL can be involved in immune response to viral infection largely through regulating TNF-α, IFN-β, IL8 expression and inflammatory response.62 Tumor Necrosis Factor (TNF), an activator of NF-κB signaling pathway, is a major inflammatory cytokine regulator in host defense against viral infection.63 THRIL directly modulates TNF-α, whereas THRIL induces other cytokines and chemokines, but the processes need to be further investigated.25 For the first time in this study, we explored the expression pattern of lncRNA THRIL in COVID-19. Similar to the expression level of MALAT-1, THRIL was significantly overexpressed in acute COVID-19 group compared to healthy samples. Comparison between acute and post-acute COVID-19 groups, the mean expression level of THRIL was significantly down-regulated during acute to post-acute phase. As well, the AUC value shows that PBMC THRIL can serve as a biomarker in the discrimination of COVID-19 patients from healthy subjects and those in the acute phase of COVID-19 from those in the post-acute phase of COVID-19 disease.
MiR-155-5p has been known as the ‘master of inflammation’ during COVID-19 disease and constitutes part of an immunopathological picture in COVID-19 disease. In inflammatory responses, miR-155-5p controls NF-κB signaling and plays a critical role in the modulating the immune response.49,64 The miR-155-5p expression is considered to be the initial step in the NF‐κB signaling upregulation of the immune cascade and feeding back through the IKK signalosome complex and PI3K/Akt to further increase NF‐κB. It has been reported that regulation of miR-155-5p levels by glucocorticoids, can be considered as one of the effective COVID-19 treatments.65 Although the expression level of miR-155-5p was upregulated in COVID-19 patients, there is no literature so far investigating miRNA-155 mechanism in COVID-19.48,52,66 However, preceding reports has demonstrated that miRNA-155 has a strong impact in NF-κB signaling.64 Our results are in line with previous studies in which the mean expression level of miR-150-5p in acute-COVID-19 subjects are significantly higher than in the healthy subjects and in post-acute COVID-19 group. In addition, according to ROC curve results for miR-150-5p, this miRNA may be considered a novel biomarker for acute COVID-19 disease diagnosis.
ConclusionAccording to the findings of the present study, expression pattern of inflammation-related ncRNAs including MALAT-1, NEAT-1, and miR-150-5p were significantly different between COVID-19 patients and healthy subjects. In addition, the level of miR-150-5p, MALAT-1, and NEAT-1 were significantly downregulated from the acute phase of COVID-19 to the post-acute phase of COVID-19. Aberrant expression of ncRNAs was found in COVID-19 disease, which maybe associated with the pathogenesis of SARS-CoV-2 and identification of these factors can be helpful in setting a basis for classification of disease conditions and acting as biomarkers and even be considered as a valuable therapeutic target for the treatment of COVID-19 diseases. Finally, the low number of samples in this study was the main limitation of our work. Hence, the assessment of these non-coding RNAs in a large population is needed.
The authors of the current survey would like to thank all of the volunteers who have participated in this research. The present study was funded by Research Deputy of Iran University of Medical Sciences, Tehran, Iran with Grant number 20975.