Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is the cause of Coronavirus Disease 2019 (COVID-19). Although Real Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR) of respiratory specimens is the gold standard test for detection of SARS-CoV-2 infection, collecting nasopharyngeal swabs causes discomfort to patients and may represent considerable risk for healthcare workers. The use of saliva as a diagnostic sample has several advantages.
ObjectivesThe aim of this study was to validate the use of saliva as a biological sample for diagnosis of COVID-19.
MethodsThis study was conducted at Infectious Diseases Research Laboratory (LAPI), in Salvador, Brazil. Participants presenting with signs/symptoms suggesting SARS-CoV-2 infection underwent a nasopharyngeal swab (NPS) and/or oropharyngeal swab (OPS), and saliva collection. Saliva samples were diluted in PBS, followed by RNA isolation and RT-Real Time PCR for SARS-CoV-2. Results of conventional vs saliva samples testing were compared. Statistical analyses were performed using Statistical Package for the Social Sciences software (SPSS) version 18.0.
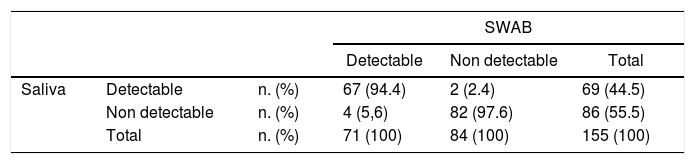
ResultsOne hundred fifty-five participants were recruited and samples pairs of NPS/OPS and saliva were collected. The sensitivity and specificity of RT-PCR using saliva samples were 94.4% (95% CI 86.4–97.8) and 97.62% (95% CI 91.7–99.3), respectively. There was an overall high agreement (96.1%) between the two tests.
ConclusionsUse of self-collected saliva samples is an easy, convenient, and low-cost alternative to conventional NP swab-based molecular tests. These results may allow a broader use of molecular tests for management of COVID19 pandemic, especially in resources-limited settings.
In December 2019, China reported the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) as the cause of Coronavirus Disease 2019 (COVID-19).1 Although Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV)2 and Middle East Respiratory Syndrome Coronavirus (MERS-CoV)3 infections have higher mortality rates than COVID-19, SARS-CoV-2 spreads much more rapidly than MERS-CoV and SARS-CoV. On March 11, 2020, the World Health Organization (WHO) announced that epidemic of the novel coronavirus was a pandemic, and as of July 8, 2020, the number of confirmed cases globally was 11,874,226, associated to over 545,481 deaths in 216 countries or territories.4 At that time, Brazil confirmed over 1.7 million cases, 67,964 deaths, and a mortality rate of 32.3/100,000.5
Direct human-to-human transmission of SARS-CoV-2 occurs through droplet inhalation while coughing, sneezing, or even talking, and transmissions caused by contact with nasal, ocular and oral mucous membranes are also possible. Several clinical symptoms are associated to COVID-19, such as fever, cough, shortness of breath, sore throat, chest pain, headache, anosmia and ageusia.6 Although 80% of cases has light or mild symptoms, the remaining 20% can present with severe disease, and around 5% will require intensive care treatment, especially those with chronic health conditions including cardiopathy, arterial hypertension, diabetes and obesity.7
Real Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR) of respiratory specimens (oropharyngeal and nasopharyngeal swabs, bronchoalveolar lavage, tracheal aspirate) is the gold standard test for detection of SARS-CoV-2 infection.8–10 However, collecting nasopharyngeal swabs causes discomfort to patients due to invasiveness of the procedure, which can reduce the possibility of patient consent to retest, and may represent considerable risk for healthcare workers, because of its potential to induce patients to sneeze or cough, expelling virus particles.11
Previous studies have shown high detection rate using saliva as specimens for laboratory diagnosis of respiratory viruses.12,13 Recent studies reported the use of oral fluids/saliva for the detection of SARS-CoV-2.14–18 The use of saliva as a diagnostic sample has several advantages, such as easy self-collection even at home, and no need of specialized personnel for sample collection. In addition, saliva collection is much more comfortable for the patient than nasopharyngeal/ oropharyngeal swabs procedure.17 It also saves time, and is less costly, because it does not require the use of personal protective equipment nor viral transportation solution.
Timely, accurate and non-invasive samples for SARS-CoV-2 detection will facilitate effective large-scale pandemic control measures to prevent the spread of COVID-19. The aim of this study was to validate the use of saliva as a biological sample for the diagnosis of COVID-19.
Material and methodsStudy populationThis study was conducted at Infectious Diseases Research Laboratory (LAPI), of Complexo Hospitalar Professor Edgard Santos (C-HUPES), Federal University of Bahia, in Salvador, Brazil.
C-HUPES healthcare workers presenting with signs/symptoms suggesting SARS-CoV-2 infection, as well as patients at the COVID-19 ward of C-HUPES, underwent a nasopharyngeal swab (NPS) and/or oropharyngeal swab (OPS) collection. All participants recruited to this study, after signing informed consent, were asked to provide a sample of self-collected saliva.
This study was approved by the Research Ethics Committee of Maternidade Climério de Oliveira – UFBA (4.042.620).
Sample collectionSaliva samples were collected into 30 ml sterile urine cups. Participants were instructed to repeatedly spit until approximately 2 ml of sample was obtained, thus avoiding mucous secretions from oropharynx or lower respiratory tract (i.e., sputum). Samples were transported to LAPI in a thermal box at 2−8 °C, and stored at -80 °C until nucleic acid extraction. Whenever possible, RNA was isolated from fresh saliva within six hours after collection.
Sample processing and viral nucleic acid isolationSamples were homogenized by repeated pipetting and diluted 1:1 with PBS 1x (phosphate buffered saline) before RNA isolation. RNA isolation was performed by using QIAGEN QIAamp® RNA Mini Kit, according to manufacturer’s instructions. Viral nucleic acid was extracted from 140 μl diluted saliva, and eluted to 60 μl of elution buffer.
Testing of samples pools with one positive and four negative samples and one positive and nine negative samples was also performed. It included positive samples (RT-Real Time PCR) with low viral load and high viral load.
RT-Real time PCRNasopharyngeal and oropharyngeal specimens were sent to Bahia Central Laboratory (LACEN), a State´s Reference Laboratory, for SARS-CoV-2 investigation by RT-PCR method, in accordance to BIOMOL OneStep/ COVID-19 Kit (Paraná Molecular Biology Institute) protocol.
For saliva samples RNA template was subjected to amplification according to Charité-Berlin protocol.10 This protocol consisted of three stages: first (screening) to amplify the E gene, a confirmatory and eliminatory step. The last two stages, targeted RdRp gene, were run in case nucleic acid was detected on screening. Amplification reactions were carried out on Applied Biosystems 7500 Real Time PCR detector, and results were classified as positive for SARS-CoV-2 when both E and RdRp genes were detected and cycle threshold (Ct, number of cycles required for the fluorescent signal to exceed background level) values were less than or equal to 40.
Immunological assayIn order to confirm the absence of SARS-CoV-2 infection in some discordant results, Enzyme Linked Immunosorbent Assay (Euroimmun, Lüebeck, Germany) was performed. Detection of IgG antibodies was attempted in plasma samples collected after three weeks of symptoms´ onset, according to manufacturer’s instructions.
Statistical analysisDescriptive statistics were presented as a percentage (%) for categorical variables and median (interquartile range; IQR) for continuous variables. Fisher’s and Mann-Whitney tests were used for categorical and continuous variables, respectively. Sensitivity, specificity, positive predictive value, negative predictive value and a 95% confidence intervals (CI) were calculated to access diagnostic performance. The kappa coefficient was used to estimate the agreement beyond chance between saliva and NPS/OPS RT-PCR results. All statistical analyses were performed using Statistical Package for the Social Sciences software (SPSS) version 18.0.
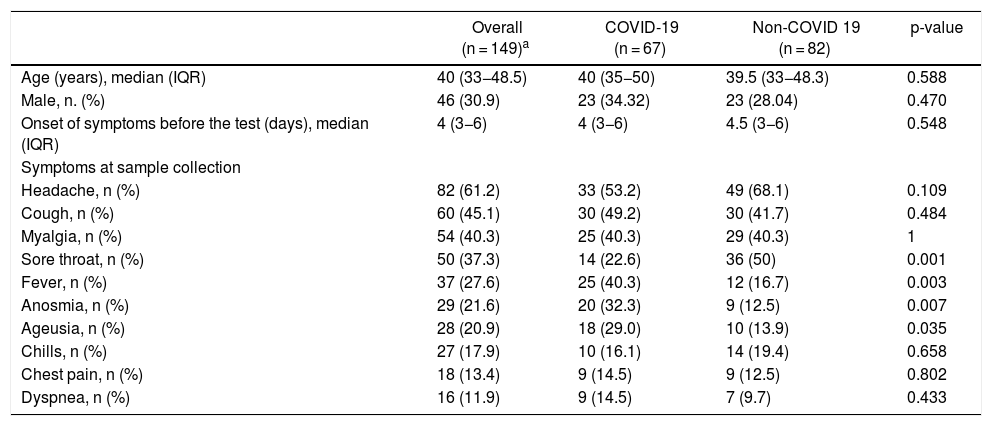
ResultsBetween May 5th and June 5th 2020, 155 participants were recruited and samples pairs of NPS/OPS and saliva were collected. Of these, 149 (96.1%) had concordant results on the detection of SARS-CoV-2 RT-PCR in both specimens. Forty-six individuals (30.9%) were male, median (IQR) age was 40 (33−48.5) years, and median (IQR) time from onset of symptoms was 4 (3−6) days. Participants´ characteristics are shown in Table 1.
Characteristics of participants enrolled in this study.
| Overall (n = 149)a | COVID-19 (n = 67) | Non-COVID 19 (n = 82) | p-value | |
|---|---|---|---|---|
| Age (years), median (IQR) | 40 (33−48.5) | 40 (35−50) | 39.5 (33−48.3) | 0.588 |
| Male, n. (%) | 46 (30.9) | 23 (34.32) | 23 (28.04) | 0.470 |
| Onset of symptoms before the test (days), median (IQR) | 4 (3−6) | 4 (3−6) | 4.5 (3−6) | 0.548 |
| Symptoms at sample collection | ||||
| Headache, n (%) | 82 (61.2) | 33 (53.2) | 49 (68.1) | 0.109 |
| Cough, n (%) | 60 (45.1) | 30 (49.2) | 30 (41.7) | 0.484 |
| Myalgia, n (%) | 54 (40.3) | 25 (40.3) | 29 (40.3) | 1 |
| Sore throat, n (%) | 50 (37.3) | 14 (22.6) | 36 (50) | 0.001 |
| Fever, n (%) | 37 (27.6) | 25 (40.3) | 12 (16.7) | 0.003 |
| Anosmia, n (%) | 29 (21.6) | 20 (32.3) | 9 (12.5) | 0.007 |
| Ageusia, n (%) | 28 (20.9) | 18 (29.0) | 10 (13.9) | 0.035 |
| Chills, n (%) | 27 (17.9) | 10 (16.1) | 14 (19.4) | 0.658 |
| Chest pain, n (%) | 18 (13.4) | 9 (14.5) | 9 (12.5) | 0.802 |
| Dyspnea, n (%) | 16 (11.9) | 9 (14.5) | 7 (9.7) | 0.433 |
The prevalence of COVID-19 diagnosed by NPS/OPS RT-PCR and by saliva RT-PCR in this study were 45.8% and 43.22%, respectively. Most of the tested healthcare workers (51.6%) were nurses/nursing technicians.
All the 67 participants diagnosed with COVID-19 had mild to moderate symptoms. No one needed hospitalization or intubation. No death was reported. The most common symptoms at sampling time were headache (53.2%), cough (49.2%) and myalgia (40.3%). Frequency of fever and anosmia was significantly higher (p = 0.003; 0.007), among patients tested positive for COVID-19, while, frequency of sore throat was significantly lower (p = 0.001) (Table 1).
Using RT-PCR of NPS/OPS samples as the gold standard, the sensitivity and specificity of RT-PCR using saliva samples were 94.4% (95% CI 86.4–97.8) and 97.62% (95% CI 91.7 – 99.3), respectively (Table 2). Positive predictive value and negative predictive value were 97.1% (95% CI 90.0–99.2) and 95.35% (95% CI 88.6–98.2), respectively. There was an overall high agreement (96.1%) between the two tests (kappa coefficient 0.922, 95% CI 0.765–1.00, p < 0.001). The median (IQR) Ct values of the E gene was 33 (29–36.6), and of the RdRp gene was 34 (29.5–37.4).
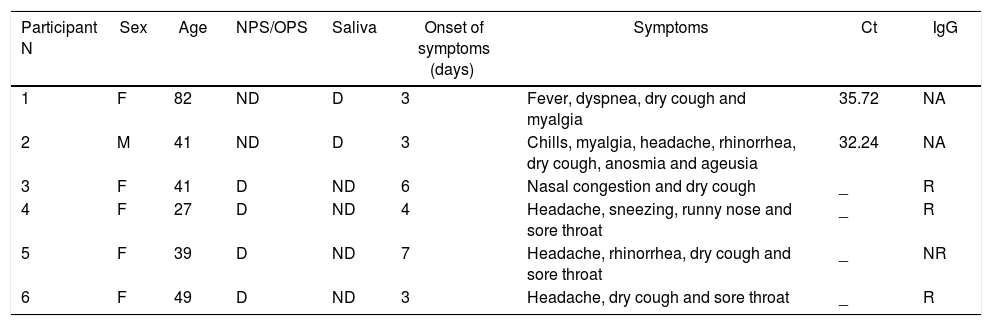
The detection rate of SARS-CoV-2 in saliva was slightly lower than that of NPS/OPS, but without reaching statistical significance. Six participants had discordant results between saliva and NPS/OPS RT-PCR assays, including two participants with virus detected in saliva but not in NPS/OPS, and four participants with virus detected in NPS/OPS but not in saliva (Table 3).
Characteristics of the participants who had discordant result on detection of SARS-CoV-2 RT-PCR by NPS/OPS and saliva.
| Participant N | Sex | Age | NPS/OPS | Saliva | Onset of symptoms (days) | Symptoms | Ct | IgG |
|---|---|---|---|---|---|---|---|---|
| 1 | F | 82 | ND | D | 3 | Fever, dyspnea, dry cough and myalgia | 35.72 | NA |
| 2 | M | 41 | ND | D | 3 | Chills, myalgia, headache, rhinorrhea, dry cough, anosmia and ageusia | 32.24 | NA |
| 3 | F | 41 | D | ND | 6 | Nasal congestion and dry cough | _ | R |
| 4 | F | 27 | D | ND | 4 | Headache, sneezing, runny nose and sore throat | _ | R |
| 5 | F | 39 | D | ND | 7 | Headache, rhinorrhea, dry cough and sore throat | _ | NR |
| 6 | F | 49 | D | ND | 3 | Headache, dry cough and sore throat | _ | R |
Abbreviations: F, female; M, male; D, detected; ND, not detected; NPS, nasopharyngeal swab; OPS, oropharyngeal swab; Ct, cycle threshold; R, reactive; NR, not reactive; NA, not available.
After 20 days of symptoms onset, participants with negative saliva results (n = 4) were invited to collect new blood samples for anti-SARS-CoV-2 antibodies. IgG was reactive in all but one of them, confirming that at least one patient probably had not been previously infected by SARS-CoV-2. Participant no 5 (negative antibody detection) had unspecific symptoms, common to any viral infection (Table 3). The two participants with saliva positive/NP-negative testing did not provide blood samples for antibody testing.
One positive and four negative (low dilution) and one positive and nine negative (high dilution) saliva samples were pooled. Low and high dilution were positive for low Ct samples (high viral load), but only positive in low dilution when a low viral load sample was included.
DiscussionOur findings demonstrate that testing saliva as an alternative to NP swabs is sensitive and specific enough to be used in a routine practice. Concordance between RT-PCR results for SARS-CoV-2 detection in saliva and NP swabs was 96.1%. In addition, among samples with discrepant results we had one that was negative for SARS-CoV-2 antibodies three weeks after diagnosis, suggesting that patient had a diagnosis other than COVID-19. In this case, the sensitivity of saliva testing would increase from 94.4 to 95.7%, while the specificity of NP swabs testing would decrease in a similar way.
The use of NP swabs has been the standard of care for SARS-CoV-2 diagnosis in Brazil, but it requires the use of full personal protective equipment, increase the risk of infection for the health professional who collects the sample, and also, the cost of testing. Saliva has been shown to be a reliable specimen for detecting SARS coronavirus since 2004. Wang et al. (2004) reported a high viral load of SARS-CoV in saliva samples when compared to throat wash.13 To et al. reported a high concordance between saliva and nasopharyngeal aspirates for the detection of respiratory viruses and monitoring SARS-CoV-2 viral load during the course of infection.12,16 There are several advantages of using saliva samples for detection of SARS-Cov-2: first, saliva can be provided easily by the patient without any invasive procedures. Therefore, the use of saliva samples could reduce the risk of virus transmission to healthcare professionals.17 Second, the use of saliva will allow sample collection outside hospitals or health centers areas. Finally, it could be conveniently pooled to screen larger populations, as demonstrated in this work. In these settings, where a large number of individuals require screening, saliva represents a practical and non-invasive specimen type.19
In the present study saliva was self-collected by participants inside the hospital area. Despite the recommendation of some authors to use samples collected immediately upon waking up,11,16 without previous food consumption or teeth brushing, we collected saliva at any time from patients who had a test requested by a physician. This easy to perform, simple procedure, provided a similar sensitivity/specificity to samples collected by NP swabs.11
In some previous studies, saliva was obtained using special collection devices,20 improving quality and quantity of obtained saliva. However, these collection devices are not usually available in general medical care centers, especially in low-income countries. Furthermore, saliva collection using such devices requires the assistance of healthcare workers. In the present study, participants were instructed to repeatedly spit into a sterile urine cup used routinely in our hospital. As no special device is required, the use of saliva can be implemented as a routine in clinical practice without compromising the sample quality, as previously reported.19,21
The high level of concordance between the two methods demonstrates that the use saliva for detecting SARS-CoV-2 by RT-PCR is as reliable as the use of NP/OP swabs, but with lower costs and risks. Brazil has almost 1.5 million cases diagnosed so far, which means that a simpler testing procedure could potentially reduce the risks and costs involved in sample collection. Despite these numbers, the proportion of patients tested by PCR in Brazil is very low, and the estimates on the dynamics of the pandemic is largely based on flow lateral immunochromatographic tests, which have been considered inaccurate to define serological status for SARS-CoV-2.22
Our results were comparable to similar studies that used saliva for SARS-CoV-2 detection and were obtained from a population of patients closely followed up in a university hospital.23 The two participants presenting negative results in saliva samples but positive in NP swabs had very non-specific symptoms and could not fulfill a clinical diagnosis of COVID-19. The most predictive symptoms for COVID-19 in our population were not present in these two cases, increasing the possibility of a false-positive result by NP swab. The use of a three-step amplification protocol also increases the sensitivity of the test. A recent study showed that some variations in SARS-CoV-2 genome can negatively impact the sensitivity of RT-PCR test used for its detection, and reinforces the need of protocols able to maintain sensitivity and specificity,24 like the one used in this study.
Our work presents some limitations including the absence of serological confirmation of COVID-19 in all cases, and the lack of ct values for the tests using NPS samples, as they were performed in a referral laboratory and were not available for comparison purposes. However, we were able to closely evaluate health care workers and inpatients admitted to our hospital and compare the use of two approaches for molecular detection of SARS-CoV-2, with a high degree of concordance.
In conclusion, we demonstrated that using self-collected saliva samples is an easy, convenient, and low-cost alternative to conventional NP swab-based molecular tests. These results can allow a broader use of molecular tests for management of COVID19 pandemic, especially in resources-limited settings.
Conflict of interestThe author declares no conflicts of interest.