The presence of Acinetobacter baumannii outside hospitals remains unclear. This study aimed to determine the prevalence of multidrug-resistance (MDR) A. baumannii in the extra-hospital environment in Mthatha, South Africa and to investigate the frequency of carbapenemase-encoding genes.
Material and MethodsFrom August 2016 to July 2017 a total of 598 abattoir samples and 689 aquatic samples were collected and analyzed presumptively by cultural methods for the presence of A. baumannii using CHROMagar™ Acinetobacter medium. Species identification was performed by autoSCAN-4 (Dade Behring Inc., IL) and confirmed by the detection of their intrinsic blaOXA-51 gene. Confirmed MDR A. baumannii isolates were screened for the presence of carbapenemase-encoding genes, ISAba1 insertion sequence and integrase intI1.
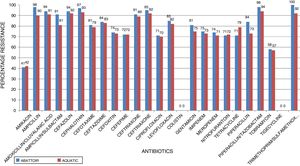
ResultsIn total, 248 (19.3%) Acinetobacter species were isolated. Acinetobacter. baumannii was detected in 183 (73.8%) of which 85 (46.4%) and 98 (53.6%) were recovered from abattoir and aquatic respectively. MDR A. baumannii was detected in 56.5% (48/85) abattoir isolates and 53.1% (52/98) aquatic isolates. Isolates showed high resistance to antimicrobials most frequently used to treat Acinetobacter infections such as piperacillin/tazobactam; abattoir (98% of isolates resistant), aquatic (94% of isolates resistant), ceftazidime (84%, 83%), ciprofloxacin (71%, 70%), amikacin (41%, 42%), imipenem (75%, 73%), and meropenem (74%, 71%). All the isolates were susceptible to tigecycline and colistin. All the isolates carried blaOXA-51-like. The blaOXA-23 was detected in 32 (66.7%) abattoir isolates and 11 (21.2%) aquatic isolates. The blaOXA-58-like was positive in 7 (14.6%) and 4 (7.7%) abattoir and aquatic isolates, respectively. Both groups of isolates lacked blaOXA-24-like, blaIMP-type, blaVIM-type, blaNDM-1,blaSIM, blaAmpC, ISAba1 and inI1. Isolates showed high level of Multiple Antibiotic Resistance Index (MARI) ranging from 0.20-0.52.
ConclusionExtra-hospital sources such as abattoir and aquatic environments may be a vehicle of spread of MDR A. baumannii strains in the community and hospital settings.
Acinetobacter baumannii has emerged over the few last decades as a cause of healthcare-associated infections with this organism associated with increased patient morbidity, mortality, and treatment costs.1–3 Its clinical significance has been propelled by its remarkable ability to upregulate or acquire resistance determinant making it one of the organisms threatening the current antibiotic era.4,5 The possession of this array of resistance mechanisms have made this organism to be able to resist almost all available antibiotics in hospital environment and has position the organism as important candidate for the evaluation of reservoirs of antibiotic resistance in the environment or even in human subjects.6Acinetobacter. baumannii have the ability to survive in both moist and dry conditions for many weeks or even months, hence this organism is widely distributed in natural and nosocomial environments, human skin, as well as mucosal surfaces facilitating its spread.7–9 The cross-transmission of this organism from patient to patient and the possibility of outbreak extension by patient transfer have been demonstrated.10 Despite hospital ecology being intensively studied, its ecology outside the hospital remains unclear or poorly defined impeding effective prevention of transmission.2,11 Several investigators have suspected that the survival of A. baumannii in the environment (in particular in water) could contribute to the transmission of the organism during outbreaks.12 It is well established that members of the genus Acinetobacter is ubiquitous but difficulty in unequivocally identify this organism has made the ubiquity in nature of A. baumannii a widespread misconception.1,9 Therefore the existence of an extra-hospital reservoir of A. baumannii and the implication of this potential reservoir in the occurrence of certain infections due to this organism are still controversial.1,2 Additionally, the prevalence and antimicrobial resistance of A. baumannii outside the hospital setting has been poorly investigated.13 There seems to be a continuous influx of novel strains into the clinical setting with the potential of new infectious features.11 Studies have shown that infections caused by extra-hospital strains of A. baumannii are responsible for community-acquired infections mainly in tropical and subtropical areas.5,14 This suggests a source of this pathogen outside of the hospital and therefore the significance of environmental isolates in the epidemiology of A. baumannii is of a great concern worldwide. There is no clear evidence about the way of introduction of A. baumannii into hospital environment, its propagation from hospital settings to the natural environment and its natural habitat outside hospitals.15 Also unlike clinical strains, there are limited reports using molecular analysis to explore the extra-hospital epidemiology of A. baumannii.11,16 The presence of A. baumannii in the aquatic environment raises the issue of public health risk since people are in constant contact with the aquatic environment.17 Hospital waste water is discharged into municipal sewage system without prior treatment, contributing to widespread contamination of natural waters with emerging pathogenic bacteria that can carry MDR genes.18 In the aquatic environment, bacteria interact with various organisms, some of which are fish used for consumption as well as commercial or recreational purposes.19 Most of the antibiotics used in the hospitals end up contaminating our local dams. The indiscriminate use of antibiotics in livestock production as growth promoters or to control infectious diseases and the treatments have been linked with the dissemination of resistant bacteria which can be transferred to people via food, direct contact with infected animal or through the abattoir. In a study conducted in Scotland, pigs and cattle slaughtered for human consumption and coming from different farms were sampled (faeces, skin, nostril and ear) for A. baumannii isolation. The prevalence of A. baumannii carriage was 1.2%. The 16 A. baumannii isolated were grouped into three different clusters, but had different pulsed-field gel electrophoresis (PFGE) patterns compared with human isolates of the three major European clones Eci, ECII, and ECIII.20 Recent reports have also described the presence of carbapenem-resistant A. baumannii in animals. The OXA-23 carbapenemase has been found in Acinetobacter from cattle, horses, and cats.21–23 The NDM-1 has also been reported in Acinetobacter spp. from food animals (chicken and pig farms) in China.24,25 However, knowledge about carbapenemase-producing A. baumannii of environmental origin (aquatic and abattoir in case of this study) remains very limited, making it difficult to assess its impact on public health. In South Africa, A. baumannii has been isolated from hospital environments; nevertheless only few studies have reported the presence of A. baumannii in the aquatic environment, meat of food animals and the environment where these meats come from. These areas are a source of water and food and therefore can be of transmission of this organism into hospital settings or may horizontally transfer resistance genes to other organisms.
Study objectivesBased on these premises, the present study was carried out to determine the prevalence of MDR A. baumannii in aquatic and abattoir environments, their susceptibility to antimicrobials and to determine the frequency of carbapenemase-encoding genes.
Material and methodsStudy design and settingsThis was a Cross-sectional prospective descriptive study conducted between the period of August 2016 and July 2017 at:
- 1
The Umzikantu red meat abattoir, a private abattoir located in South Ridge Park, O.R. Tambo district municipality in Mthatha, Eastern Cape. This red meat abattoir is a type E5 abattoir. It operates as an abattoir and meat wholesalers and it is the only operational abattoir in Mthatha. Abattoirs differ greatly in the scale of their operations. Using the EU definition of livestock units, the scale is graded according to the number of beast they are designed to handle (without some health risk) per week as follows: E: - to slaughter five units, D: - to slaughter 15 units, C: - to slaughter 50 units, B: - to slaughter 200 units, and A: - to slaughter 1000 units (exporting type). In its basic form one EU Livestock Unit (ELU) equals: one cow beast, two calves, five pigs or 10 sheep.26
- 2
The Mthatha dam (coordinates: 31°33′2″S 28°44′24″E), an earth-fill type dam which impounds the Mthatha river at a location about eight kilometers upstream of the city of Mthatha in the OR Tambo District Municipality of the Eastern Cape Province. The dam was established in 1977 and serves mainly for municipal and industrial purposes. Its hazard potential has been ranked high. The storage capacity of the Dam is 253,674,000 cubic meters. It serves as the main source of raw water supply (65.9 mL/day) for the Thomhil WTW which treats and supplies water to the city and its environs with a total population of about 750,000.
Five hundred and ninety-eight samples were recovered from the hides, from structural and work surfaces (equipment, floors, doors, knives, saws, handling panels from the slaughter area) within the abattoir and from carcasses and meat at all stages of processing (from flaying to evisceration, splitting and cooling). At each site, an area of 10 cm2 was sampled using a sterile swab moistened with physiological saline. Animal samples were also collected from recently slaughtered animals destined for human consumption. Carcasses were sampled by systematic random sampling technique. Swabs were taken according to the method described by the international organization for standardization.27 The abdomen (flank), thorax (lateral), crutch, breast (lateral), were the sampling sites. Swabbing was performed approximately six hours after cleaning and disinfection and prior to slaughter activities in the abattoir. The samples were cooled and transported to the laboratory for plating within 10 h after sampling. For meat samples, 25 g of meat were cut aseptically into very small pieces and homogenized by using a stomacher bag (Interscience, Saint Nom, France) and then suspended in sterile water.
Sample collection from aquatic environmentOverall, 689 samples were collected from the Mthatha dam. The dam was sampled five times within the study period and samples were collected using aseptic techniques in the mornings (between 08:00 and 10:00 h local time). Duplicate water samples were collected at margins and in the middle of the dam, about 15 cm down the water surface in 100 ml sterilized bottles. The collected water samples were placed in a cooler box with temperature maintained between 4 and 10⁰C using ice packs. These samples were immediately transported to the Walter Sisulu University laboratory and subjected to serial ten-fold dilutions in sterile distilled water. Forty-nine fishes belonging to different species were collected aseptically and immediately transported in a thermal bag to the laboratory where they were sacrificed. Samples from skin, gills, intestines, and eyes were collected. The body surface and gills were carefully washed, at first under the stream of tap water and then using 70% ethyl alcohol to remove normal external bacterial flora. Then, the fish were disinfected with 70% ethanol and after opening the body cavity, samples from the kidneys and intestine were taken. The samples were diluted 1:1 in PBS and homogenized.
Bacterial cultivation and identification
Samples were presumptively identified by direct plating on Chromogenic CHROMagar™ Acinetobacter (CHROMagar™; Paris, France) with supplement Ref. CR102 which allows the growth of carbapenem-resistant isolates of Acinetobacter species, incubated at 37 °C in aerobic conditions and examined after 24 h for the growth of typical red colonies of Acinetobacter species. CHROMagar™ Acinetobacter was prepared according to the manufacturer’s description. Cefsulodin sodium salt hydrate (Sigma-Aldrich) was added at 15 mg/L to suppress the growth of Pseudomonas and Aeromonas species. Gram-positive bacteria and yeasts are inhibited. Confirmation of presumptive colonies was performed by inspection of colony morphology on the separate plates. The plates were examined for the growth of typical red colonies of Acinetobacter species. One typical colony representative of each type of morphology and shape was further sub-cultured on tripticase soy agar (TSA) and characterized based on phenotypic tests: Gram-stain, catalase and oxidase tests. Gram-negative coccobacilli, oxidase-negative, and catalase-positive isolates were presumptively identified to the genus Acinetobacter. Species identification were carried out using Gram-negative ID type 2 panel (REF. B1017-27) of the MicroScan autoSCAN-4 automated System (Dade Behring Inc., Deerfield, IL). A high percentage (≥95%) was utilized as the acceptance criterion for identification by MicroScan. Suspected colonies were also further verified using the Acinetobacter specific primer set Ac436 F and Ac676 r to amplify the 16S rRNA gene. Confirmation of A. baumannii isolates was carried out by polymerase chain reaction analysis of the presence of inherent BlaOXA-51-like genes. Isolates lacking the BlaOXA-51-like gene were investigated further by sequence analysis of the 16S rRNA gene. Primers were ordered from TIB Molbiol, Germany. All the strains were freshly suspended in skim milk (Merck, Germany) containing 15% glycerol in sterile glass, screw-capped vials and stored at -80 °C until further use.
Antimicrobial susceptibility testingAntimicrobial susceptibility testing was performed by MicroScan autoSCAN-4 System. The antibiotics tested were amikacin (AMK), amoxicillin/clavulanic acid (AMC), ampicillin/sulbactam (AMS), ampicillin (AMP), cefazolin (CEP), cefepime (FEP), cefotaxime (CTX), ceftazidime (CAZ), ceftriaxone (CRO), cefuroxime (CXM), cefoxitin (FOX), cephalothin (CEPH), ciprofloxacin (CIP), gentamicin (GEN), imipenem (IMP), meropenem (MEM), levofloxacin (LVX), nitrofurantoin (NIT), tetracycline (TEC), tobramycin (TOB), trimethoprim/sulfamethoxazole (SXT), piperacillin/tazobactam (TZP), piperacillin (PRL), colistin (CST), and tigecycline (TGC) according to the CLSI guidelines v27.28 Non-susceptibility was defined as the combination of resistance and intermediate resistance. MDR A. baumannii isolates were defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories.29Escherichia coli (ATCC #25922) and Pseudomonas aeruginosa (ATCC #27853) were included as quality control strains.
Multiple antibiotic resistance index (MARI)The MARI was calculated and interpreted according to Krumperman (1983) as the ratio of the number of antibiotics to which the isolates were resistant (a) to the total number of antibiotics to which the isolates were exposed (b), i.e: MARI = a/b.
The MARI of individual A. baumannii from the two different sources was calculated. Bacteria having MARI (>0.2) originate from a high risk source of contamination where several antibiotics are used. MARI value of ≤0.2 indicates strain originated from sources where antibiotics are seldom or never used.30
Investigation of carbapenemase-encoding genes, ISAba1 and intI1 by PCRGenomic DNA from an overnight culture on tryptic soy broth of A. baumannii was extracted using the MagNaPure Compact® nucleic acid isolation kit I (REF. 03730964001, Roche diagnostics, Mannheim, Germany) according to the manufacturer’s protocol. The presence of the genes encoding Ambler class D Serine-Carbapenemase (blaOXA-51-like,blaOXA-23-like, blaOXA-24-like, and blaOXA-58-like), Ambler class B Metallo-β-lactamases (blaIMP-1, blaVIM, blaSIM and blaNDM-1) and Ambler class C blaAmpC were analyzed by PCR. ISAba1 and intI1 were also determined. The presence of the ISAba1 upstream blaOXA-23-like and blaOXA-51-like genes to promote gene expression were performed using ISAba1 forward/blaOXA-23-like reverse or blaOXA-51-like reverse primers.31 The details of primer sequences used for PCR amplification of the resistant genes are listed in Table 1.
List of primers used for PCR amplification of resistant genes.
| Gene | Oligonucleotide sequence | Fragment size (bp) | Location | References |
|---|---|---|---|---|
| 16SrRNA | F: 5’TTT AAG CGA GGA GGA GG3’ R: 5’ATT CTA CCA TCC TCT CCC3’ | 240 | 16SrRNA | 32 |
| OXA-23-like | F: 5’GATCGGATTGGAGAACCAGA3’ R: 5’ATTTCTGACCGCATTTCCAT3’ | 501 | blaOXA-23 | 33 |
| OXA-24-like | F: 5’GGTTAGTTG GCC CCC TTA AA3’R: 5’AGTTGAGCGAAAAGGGGATT3’ | 246 | blaOXA-24 | 33 |
| OXA-51-like | F: 5’TAATGCTTT GATCGG CCT TG3’ R: 5’TGGATTGCACTT CAT CTT GG3’ | 353 | blaOXA-51 | 33 |
| OXA-58-like | F: 5’AAGTATTGGGGCTTGTGCTG-3’R: 5’CCCCTCTGCGCTCTACATAC3’ | 599 | blaOXA-58 | 33 |
| IMP-1 | F: 5’GATGGTATGGTGGCTCTTGT3’R: 5’TTAATTTGCCGGACTTAGGC3’ | 448 | BlaIMP | 34 |
| NDM-1 | F: 5’ATTAGCCGCTGCATTGAT3’ R: 5’CATGTCGAGATAGGAAGTG3’ | 154 | blaNDM | 35 |
| VIM-like | F: 5’ACTCACCCCCATGGAGTTTT3’R: 5’ACGACTGAGCGATTTGTGTG3’ | 815 | blaVIM | 36 |
| SIM-1-like | F: 5’TAATGCTTT GATCGG CCT TG3’ R: 5’TGGATTGCACTT CAT CTT GG3’ | 353 | blaSIM | 33 |
| AmpC | F: ACAGAGGAGCTAATCATGCGR: GTTCTTTTAAACCATATACC | 1243 | blaampC | 37 |
| ISAba1 | F: 5-CACGAATGCAGAAGTTG-3R: 5-CGACGAATACTATGACAC-3 | 599 | ISAba1 | 38 |
| ISAba1/blaOXA-23-like | F: AATGATTGGTGACAATGAAGR: ATTTCTGACCGCATTTCCAT | 1433 | ISAba1/blaOXA-23 | 38 |
| ISAba1/blaOXA-51-like | F: AATGATTGGTGACAATGAAGR: TGGATTGCACTTCATCTTGG | 1252 | ISAba1/blaOXA-51 | 31 |
| intI1 | F: 5’CAG TGG ACA TAA GCC TGT TC3’R: 5’CCC GAC GCA TAG ACT GTA3’ | 160 | Integrase gene (intI1) | 39 |
Ethical approval for this project was obtained from the Walter Sisulu University Research Ethics and Biosafety Committee [Reference number: 019/2016]. Also written permission was sought from the authorities of the two study sites. For the protection of animals used for scientific purposes the experiments described complied with the guidelines of the European Union Council http://ec.europa.eu/environment/chemicals/lab_animals/legislation_en.htm (Directive 2010/63/EU).
Statistical analysisThe data were analyzed using the Statistical Package for Social Sciences (SPSS, IBM, version 23.0 Armonk, NY). The frequency of some parameters including A. baumannii susceptibility and also resistant genes were compared between abattoir and aquatic A. baumannii by Chi-square or Fisher’s exact test. P-values <0.05 were considered as significant.
ResultsAntimicrobial susceptibility testing and MARIA total of 1287 samples were collected from the abattoir and aquatic sources from which 248 (19.3%) Acinetobacter species were isolated. A. baumannii was the predominant species (n = 183, 73.8%) of which 85 (46.4%) and 98 (53.6%) were recovered from abattoir and aquatic, respectively. Of these a total of 100 MDR A. baumannii isolates, including 56.5% (48/85) abattoir and 53.1% (52/98) aquatic, were selected for analysis. Table 2 shows the distribution of the identified Acinetobacter species according to the sources of samples. All MDR isolates showed a susceptible phenotype in response to colistin and tigecycline. Among abattoir isolates, all (100%) were resistant to trimethoprim-sulfamethoxazole, and >70% were resistant to cephalosporins (ceftazidime, cefazolin, cephalothin, cefotaxime, ceftriaxone, cefepime, cefuroxime, and cefoxitin), gentamycin and ciprofloxacin. Carbapenem resistant was 75% against imipenem and 74% against meropenem. However, there were considerable differences in resistance rates and patterns between the isolates recovered from the two study areas. Chi-square test revealed that there were no significant differences in resistance rates among different sources of isolates (p < 0.253). The resistance rates against imipenem, meropenem, ciprofloxacin, and gentamycin were 73%, 71%, 70%, and 75%, respectively, in isolates recovered from aquatic source, whereas resistance rates were 57–94% against other antimicrobial agents except for amikacin (42%). Fig. 1 shows the resistance characteristics of these isolates. The A. baumannii exhibited 25 antibiotic resistant patterns with high level of MARI (>0.2) ranging from 0.20–0.52.
Distribution of the identified Acinetobacter species according to the sources of samples.
| Acinetobacter species isolated | Total no. of isolates | Abattoir isolates | Aquatic isolates | ||
|---|---|---|---|---|---|
| Source | Number of isolates found | Source | Number of isolates found | ||
| MDR A. baumanniiNon-MDR A. baumannii | 10083 | FloorsDoorsKnivesSawsHandling PanelsSlaughter AreaAbdomen (Flank)Thorax (Lateral)CrutchBreast (Lateral)TotalFloorsDoorsKnivesSawsHandling PanelsSlaughter AreaAbdomen (Flank)Total | 519724732848849645137 | SedimentWater SampleGillsIntestinesEyesSkinTotalSedimentWater SampleGillsIntestinesEyesSkinTotal | 1715535752915635846 |
| Total A. baumanniistrains (MDR andNon-MDR)Acinetobacter lwoffii | 18328 | FloorsDoorsKnivesSawsHandling PanelsSlaughter AreaAbdomen (Flank)Total | 85233224117 | SedimentWater SampleGillsIntestinesEyesSkinTotal | 9832131111 |
| Acinetobacter calcoaceticus | 24 | FloorsDoorsSawsHandling Panels Slaughter AreaTotal | 3421111 | SedimentWater SampleGillsIntestinesEyesSkinTotal | 42121313 |
| Acinetobacter pittii | 13 | FloorsDoorsHandling PanelsSlaughter AreaTotal | 21115 | SedimentWater SampleIntestinesSkinTotal | 21418 |
| Total | 248 | 118 | 130 | ||
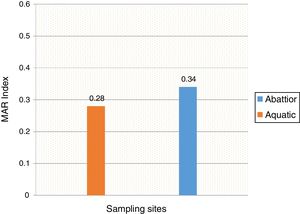
Mean MARI of isolates from both sources are shown in Fig. 2. The majority of the A. baumannii from the abattoir source (31 isolates, 64.6%) were resistant to at least eight antibiotics whilst the majority of A. baumannii from the aquatic source (21 isolates, 40.4%) were resistant to six antibiotics (Table 3).
Multiple antibiotic resistance among A. baumannii isolates from sampling sites (n = number of isolates).
| Sampling sites | Five antibiotics (n) (%) | Six antibiotics (n) (%) | Seven antibiotics (n) (%) | >Seven antibiotics (n) (%) |
|---|---|---|---|---|
| Abattoir | 2 (4.2%) | 10 (20.8%) | 5 (10.4%) | 31 (64.6%) |
| Aquatic | 7 (13.5%) | 21 (40.4%) | 6 (11.5%) | 18 (34.6%) |
The two groups of MDR isolates were analyzed for carbapenemase-encoding resistance genes (blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, blaOXA-58-like, blaIMP-1, blaSIM, blaVIM, blaampC and blaNDM-1). The blaOXA-51 is intrinsically found in A. baumannii and was detected in all abattoir and aquatic isolates which is consistent with the blaOXA-51-like gene as an intrinsic carbapenemase. A total of 32 (66.7%) of the 48 abattoir isolates carried blaOXA-23, whereas 11 (21.2%) of the 52 aquatic isolates were blaOXA-23 positive. The blaOXA-58-like was detected in seven (14.6%) and four (7.7%) abattoir and aquatic isolates respectively. There were no blaOXA-24-like, blaIMP-type, blaVIM-type, blaNDM-1, blaSIM, and blaampC genes among both groups of isolates. All isolates lacked ISAba1 and ntI1. According to Chi-square test, there were significant differences in the detection of resistant genes among different sources of isolates (p < 0.001).
DiscussionOver the past two decades, Acinetobacter species have become virulent pathogens, responsible for nosocomial infections and outbreaks, particularly in intensive care units and high dependency units (HDUs).2 Majority of published studies have concentrated on the hospital epidemiology of these organisms2 and animal health care settings making it difficult to demonstrate the extra-hospital origin of A. baumannii. The intensive use of antibiotics over the last few decades in human and as growth promoting and as prophylactic agents in livestock have resulted in serious environmental and public health problems, since this enhances antimicrobial selective pressure. Also anthropological activities have great impacts on our local dams. The antibiotic residues concentration increases in hospital and industrial effluents and these end up in our local dams leading to the development of antibiotic-resistant bacteria. Gros et al., studied the antibiotic concentrations in aquatic environments and the median concentrations in surface and ground water were reported as 0.030 and 0.071 µgL−1, respectively.40 These concentrations are above the MIC/MBC for all antibiotics, according to the British Society for Antimicrobial Chemotherapy.41A. baumannii isolated from various environmental locations has been linked with nosocomial spread.42 The resistant bacteria from the extra-hospital environments may be transmitted to humans, in whom they cause disease that cannot be treated by conventional antibiotics. In aquatic environment two hypotheses can arise from the detection of A. baumannii: (1) water is a normal habitat of A. baumannii, or (2) the presence of the bacterium results from human or animal contamination. To our knowledge, this is the first study investigating the molecular mechanisms accounting for the multidrug-resistance observed in A. baumannii recovered from abattoir and aquatic samples in the Eastern Cape Province of South Africa.
In the current study, the prevalence of A. baumannii was 14.2% in abattoir and 14.0% in aquatic samples collected. Of these, MDR A. baumannii was recovered from 56.5% (48/85) abattoir isolates and 53.1% (52/98) aquatic isolates and were analyzed. Therefore, our study evidenced the extra-hospital presence of A. baumannii in Eastern Cape Province of South Africa. This is in agreement with a previous study that found 51.2% (85/166) from meat samples to be MDR,9 i.e. acquired resistance to at least one agent in three or more antimicrobial categories among the eight different classes of antibiotics used to define MDR.29 In contrast, a study conducted by Stenstrom et al. in Nkonkobe Municipality of the Eastern Cape Province, South Africa, found that the prevalence of Acinetobacter calcoaceticus complex from soil and water samples in Alice town was 41%.6 Also previous studies in Senegal, Scotland, and La Reunion Island isolated A. baumannii as animal colonizers in 5.1%, 1.2%, and 6.5%, respectively.5,13,20 The study also revealed the quantitative changes in antibiotic resistance at two sampling sites. In general, susceptibility testing showed that 98% in abattoir isolates and 90% in aquatic isolates were resistant to ampicillin which is usually a non-effective antibiotic for the treatment of Acinetobacter infections. A high incidence of ampicillin resistance in aquatic environments was reported.43 High resistance to trimethoprim/sulfamethoxazole in abattoir (100%) and aquatic isolates (92%) is a result of the inherent resistant of A. baumannii to this compound through the target protein, dihydrofolate reductase, which has a low affinity for the drug44 and also to selective pressure caused by the use of these antimicrobial agent for treatment of infections in cattle. The MARI is a good risk assessment tool and its value (nominally 0.20) has been applied to differentiate low- and high-risk regions where antibiotics are overused.45 The present study showed MARI ranging from 0.20 to 0.52 (mean MARI; aquatic, 0.28 and abattoir, 0.34) which indicates that the isolates emerged from high-risk sources of contamination and also gives an idea of the number of A. baumannii isolates showing antibiotic resistance in the risk zone of the susceptibility study. This indicates that the study areas where these isolates came from were exposed to an environment of high-risk contamination from a region or area where there is high antibiotic use,30 probably from hospital and industrial effluents which find their way into aquatic environment or the indiscriminate use of antibiotics in the livestock slaughtered at the abattoir. This can lead to high antibiotic resistance selective pressure.46 The high resistance rate to ciprofloxacin, gentamycin, and tetracycline was not expected. These antimicrobials may have been used in these animals either as growth promoter, prophylactic or for treatment in case of abattoir isolates and also ending in aquatic environment. This result reflects the use of antimicrobial agents in livestock in South Africa, as tetracycline has long been the antimicrobial most commonly used in livestock, accounting for over 30% of the total amount of antimicrobial consumption.47 Furthermore the high resistant rate to cephalosporins may be because this antibiotic is frequently used in hospitals and might have found their way into the aquatic environment in case of the isolates from the dam or might have been introduced as a growth promoter, prophylactically or for treatment of food animals that were slaughtered at the abattoir. The current study showed that all isolates from both sources were susceptible to colistin and tigecycline which are last line antibiotics. Colistin and tigecycline are normally used as a drug of last resort in hospitals which limits the possibility of bacteria developing resistance against these antibiotics. Also, they are rarely used in animals either as a growth promoter, prophylactically or for treatment. It is also noteworthy to highlight the fact that there was no statistical difference in the resistance rates of A. baumannii isolates recovered from the abattoir and from the aquatic environment (p < 0.253). This wide range of resistance, as demonstrated here, is a cause for concern.
In this study, isolates were also tested molecularly for gene similarities. Molecular analysis of the 48 and 52 MDR A. baumannii isolates showed that all isolates from both sources harboured blaOXA-51–like and 32 (66.7%) and 11 (21.2%) isolates harboured blaOXA-23–like genes in abattoir and aquatic isolates, respectively. This indicates that blaOXA-23 was the most prevalent carbapenem hydrolyzing class D β-lactamase gene frequently found in carbapenem-resistant A. baumannii isolates collected from extra-hospital environments. Our study showed that blaOXA-23 producers in particular and carbapenemase producers in general, may be isolated from extra-hospital environments. Among the hypotheses that could explain the selection of this carbapenemase is the indiscriminate use of penicillins or penicillin–β-lactamase inhibitor combinations. This could create selective pressure for β-lactamases because blaOXA-23 does confer, in addition to decreased susceptibility to carbapenems, a high level of resistance to those compounds. It is well established that animals can be reservoirs of antimicrobial-resistant bacteria.48 These identified determinants are not only of concern in A. baumannii. Rather, these genes can be transferred to other bacteria by horizontal gene transfer and are of great concern to human health. Therefore, there is the need to emphasize on strict adherence to personal and general hygiene to reduce the risk of opportunistic infection by A. baumannii which is difficult to control. Cases of Acinetobacter infection have been traced to environment due to poor hygiene, especially hand hygiene.1
The current study showed that all MDR isolates lacked significant antibiotic resistance features, such class 1 integrons and ISAba1, indicating that the class 1 integron and ISAba1 associated resistance traits are not implicated in MDR of environmental A. baumannii. This is in agreement with a study conducted by Hamouda et al. where no class 1 integrons and ISAba1 were found in A. baumannii isolates from recently slaughtered animals destined for human consumption and collected at major Scottish abattoirs.20 ISAab1 is considered the first step in resistance evolution in A. baumannii.31 The absence of these resistance-driven mobile elements in these sources suggests that no or little antibiotic selective pressure was applied to these pathogens, contrary to their clinical counterpart. Therefore the high resistance rate to carbapenems which is the drug of choice to treat A. baumannii infections in both abattoir and aquatic isolates can be partly linked to the increased expression level of blaOXA-51-like carbapenemases. In addition, it is speculated that ISAba1 attains full expression under conditions of excessive selective pressure (presence of carbapenems); thus, one would not expect to find this mechanism of resistance in an environmental isolate.14 However, three isolates (AB4, AB28 and AF3) that expressed blaOXA-51-like in our study unexpectedly showed imipenem MICs that were lower than 4 μg/mL, which suggests that imipenem resistance in some A. baumannii strains may be primarily modulated by genes other than blaOXA-51-like. This can also be due to the blaOXA-51-like genes being usually silent or poorly expressed,49 thus not conferring a resistance phenotype. Also, the increased use of carbapenems antibiotics to treat food producing animals may explain the high level of resistance to antibiotics of this class observed in the current study. The present results provide evidence that extra-hospital A. baumannii isolates serve as a reservoir of multiple antibiotics resistant and hence as potential route for the entry of MDR pathogens into human population. Our results also suggest that the nosocomial pathogen A. baumannii is well adapted to different environments, not only to the hospital setting. This has very important implications for human health, as infections by MDR are difficult to treat and often requires expensive antibiotics, long term therapy and even mortality. Further studies should be performed ascertain the prevalence of MDR A. baumannii in other sources.
ConclusionThis study has demonstrated by molecular identification that MDR A. baumannii is actually present outside the hospital. The presence of MDR A. baumannii in the extra-hospital environment may be a threat to public health considering that this may provide a vector for the spread of these opportunistic pathogen into both community and hospital settings environment. Coordinated approaches to reduce integrated human health risks in the environment as well as careful compliance with the WHO guidelines50 on surveillance, rational antibiotic prescribing, and standard treatment guidelines for both community- and hospital-acquired infections will lead appreciably towards reducing the ever-rising threat of antibiotic resistance. However, further studies are needed for better understanding of the mechanism of interactions between the different potential reservoirs and humans. Also, possible impact on the horizontal transfer of blaOXA genes, surviving in selected condition or occurrence of infection outside the hospital setting should be further investigated.
FundingFunding of the research through the National Research Foundation (NRF) Thuthuka grant UID: 107619 is graciously appreciated. Beyond this, it did not take part in the research activities of knowledge creation and dissemination. The Nelson Mandela Academic Hospital (NMAH) and the Microbiology Laboratory at the National Health Laboratory Services (NHLS) at the NMAH are also acknowledged.
Availability of data and materialsAll data generated or analyzed during this study are included in this published article (and its supplementary information file).
Authors’ contributionsConceived and designed the experiments: S.S.P, T.A., S.D.V., A.Y.A. Performed the experiments: S.D.V., A.Y.A. Analysed the data: S.S.P, T.A., S.D.V, A.Y.A, O.G. E. Contributed reagents/materials/analysis tools: S.S.P, O.G. E., S.D.V Analysed data and wrote the manuscript: S.S.P, T.A., S.D.V, A.Y.A. All authors read and approved the final manuscript.
Ethical approvalEthical approval for this project was obtained from the Walter Sisulu University Research Ethics Committee (Human) [Reference number: 019/2016].
Conflict of interestsThe authors declare that there is no conflict of interests regarding the publication of this paper.
The authors warmly acknowledge the host, Walter Sisulu University and the co-host, University of KwaZulu-Natal.