Rhodococcus equi is an opportunistic pathogen, causing rhodococcosis, a condition that can be confused with tuberculosis. Often, without identifying M. tuberculosis, physicians initiate empiric treatment for tuberculosis. R. equi and M. tuberculosis have different susceptibility to drugs. Identification of R. equi is based on a variety of phenotypic, chromatographic, and genotypic characteristics.
ObjectiveThis study aimed to characterize bacterial isolates from sputum samples suggestive of R. equi.
MethodsThe phenotypic identification included biochemical assays; thin-layer chromatography (TLC) and polymerase chain reaction (PCR) were used for genotypic identification.
ResultsAmong 78 Gram-positive and partially acid-fast bacilli isolated from the sputum of tuberculosis-suspected patients, 51 were phenotypically and genotypically characterized as R. equi based on literature data. Mycolic acid analysis showed that all suspected R. equi had compounds with a retention factor (Rf) between 0.4-0.5. Genotypic characterization indicated the presence of the choE gene 959bp fragments in 51 isolates CAMP test positive. Twenty-two CAMP test negative isolates were negative for the choE gene. Five isolates presumptively identified as R. equi, CAMP test positive, were choE gene negative, and probably belonged to other bacterial species.
ConclusionsThe phenotypic and molecular techniques used constitute a good methodological tool to identify R. equi.
Rhodococcus equi is considered a bacterial agent of medical importance in the group of mycolic-acid-containing bacteria or aerobic actinomycetes. R. equi is the main species of the genus Rhodococcus causing infections (rhodococcosis) in animals and humans. This bacterial agent has emerged as a significant opportunistic pathogen in immunocompromised people, particularly, human immunodeficiency virus (HIV)-infected patients. Rhodococcosis affects mainly the lungs, with clinical and pathological features similar to pulmonary tuberculosis.1,2
The R. equi cell wall includes mycolic-acids of carbon length 30-54 atoms, resulting in a highly variable acid-fast staining. The organism presents the shape of partially acid-fast bacilli (PAFB). Because of its morphologic characteristics, R. equi can easily be mistaken for mycobacteria, and may be associated with invasive or systemic infections.2,3
Diagnosis of pulmonary rhodococcosis may be obtained from analysis of clinical specimens such as sputum, bronchial-alveolar lavage (BAL), and biopsy. Smears of clinical specimens stained by modified Kinyon or Ziehl-Neelsen methods may show PAFB.4 Culture and biochemical identification are, however, the gold standard methods for confirming the presence of the pathogen. The colonies grow satisfactorily in nonselective culture media, including those used in the isolation of mycobacteria and fungi.5
The phenotypic identification of R. equi is obtained by classical morphological and biochemical tests. Initially, a presumptive identification is determined by the equi factor production, shown by a positive CAMP test reaction.6 Biochemically, R. equi is non-reactive and does not oxidize or ferment carbohydrates in standard Gordon media, and does not use acetate, citrate, or malonate as unique carbon sources. It produces catalase and lipase.4,6
R. equi may also be presumptively identified through thin layer chromatography (TLC) by detection of compounds (mycolic acids), with a retention factor (Rf) between 0.4 and 0.5.7,8Identification of R. equi by polymerase chain reaction (PCR), a widely used molecular method, is possible through detection of the vapA gene present in an 85kb plasmid associated with virulence. However, this plasmid is not always present in isolates of human origin.9–12
Another effective PCR assay is that described by Ladrón et al.13 for species-specific R. equi identification, which is based on the amplification of a 959bp fragment of the choE gene. This is a chromosomal locus encoding cholesterol oxidase (COX), a secreted enzyme considered as a R. equi virulence factor.1,13 COX is the cytolytic factor responsible for the synergistic hemolysis that occurs in the CAMP test reaction, elicited by R. equi in the presence of sphingomyelinase C-producing bacteria, such as Listeria ivanovii, Bacillus cereus, and Staphylococcus aureus.1
The similarity of the clinical courses of rhodococcosis and tuberculosis and the high prevalence of tuberculosis in Brazil14 make it difficult to detect R. equi infection and establish an appropriate treatment. Considering the clinical importance of R. equi, this study aimed to identify the species in bacterial isolates recovered from sputum samples. A set of techniques, including TLC, is proposed for phenotypic identification, and PCR to characterize genotype, thus establishing an algorithm for the identification of R. equi.
Material and methodsBacterial isolates and microbiological proceduresSeventy-eight bacterial isolates (PAFB) suspected of Rhodococcus spp. and the reference strain R. equi ATCC 6939 were included in this study. The bacterial isolates were recovered in cultures of sputum samples obtained from patients suspected to have pulmonary tuberculosis, attending public health units in Ribeirão Preto, SP, Brazil. The phenotypic characterization was performed by the Laboratory of Bacteriology of the Instituto Adolfo Lutz, and analysis of mycolic acid and molecular identification was performed by the “Professor. Dr. Hugo David” Mycobacteria Laboratory, Pharmaceutical Sciences School, Universidade Estadual Paulista (UNESP) – Araraquara, SP, Brazil.
R. equi phenotypic identificationThe bacterial isolates were cultured on Müeller-Hinton agar (MH) plates and isolated colonies were transferred to tubes with MH agar slants. Standardized techniques, including Gram- and modified acid-fast staining plus additional tests were used to characterize Rhodococcus species in bacterial isolates.15,16 The isolates were additionally tested by the CAMP test, performed on sheep blood agar plates with MH base medium (Dfico) and Listeria ivanovii ATCC 19119 as the indicator strain. As previously described, the R. equi reference strain ATCC 6939 was included as a control.17
Analysis of mycolic acid by TLCThe analysis of mycolic acid was performed according to the technique recommended by Miyaji et al.,8 with some modifications. Bacterial isolates were plated on MH agar and the cultures were kept at room temperature until satisfactory growth was observed (up to seven days). The extraction of mycolate methyl-ester was performed in screw cap glass tubes (13 x 100 mm) containing the bacterial suspensions (± 30mg of bacteria) in one mL of methanol/toluene/sulfuric acid (30/15/1). After incubation at 75°C from 10 to 12hours to extract the methyl-ester mycolate, the tubes containing the extract received one mL of hexane at room temperature, and were vortexed and left to stand for the separation of the two phases. The supernatants containing methyl-ester mycolate were transferred (0.5mL) to other tubes, which were kept at 45°C to evaporate the hexane. To perform TLC, dry residues of methyl-ester mycolate were dissolved in 40mL of hexane and applied by capillary tubes on silica gel plates (Merck®, Silica gel 60F254). The plates were placed in a glass chamber (22 x 22 x 10 cm) containing 100mL of hexane/diethyl ether (8/2) for the chromatographic separation of the components. Sprays of 10% ethanolic solution of rhodamine were used to analyze results. The presence of mycolic-acids spots was determined by the Rf. Reference strains (Corynebacterium pseudodiphthericum IAL 0104, C. xerosis IAL 0105, Nocardia asteroides IAL 2125, N. brasiliensis IAL 2126, N. brevicatena IAL 2123, Rhodococcus equi ATCC 33701, R. equi ATCC 33702, R. equi ATCC 33703, R. equi ATCC 6939, Mycobacterium aurum ATCC 23366, M. vaccae ATCC 15483 and M. smegmatis ATCC14468) were also tested for the presence of mycolic-acids.
Identification of R. equi by PCRThe identification of R. equi by PCR was performed according to Ladrón et al.,13 with some modifications. Genomic DNA was obtained according to Telenti et al.18. Briefly, a loopful of bacteria grown on solid medium (MH agar) was suspended in 300μL of TE buffer (10mM Tris, 1mM EDTA [pH 8]) in 2.5mL tubes, boiled for 10minutes and then frozen (-70°C) for 20minutes. The boiling and freezing procedures were repeated twice. After stabilization at room temperature, the tube contents were submitted to the process of extraction and purification of DNA using the procedures established by Van Soolingen et al.19, with modifications. Briefly, 600mL of phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol) were added to the tubes, and the mixture was vortexed for 10 s. After centrifugation (10,000g at 4°C) for 20minutes, the tubes were left to stand until phase separation. The aqueous top phase containing DNA was transferred to other tubes, and mixed with 600mL of chloroform-isoamyl alcohol (24:1, vol/vol). This was followed by vortexing for 10 s, centrifugation (10,000g at room temperature) for five minutes, and standing for the separation of the phases formed. The aqueous top phase was transferred to another tube, and the procedure was repeated once more. Precipitation of DNA in the final aqueous phase was performed by addition of absolute ethanol (300mL) and centrifugation (12,000g at 4°C) for 20minutes. The supernatant was discarded and the pellet was resuspended in 500mL of 70% ethanol (previously kept at -20°C, for at least 18hours) and centrifuged (12,000g at 4°C) for 20minutes. The pellet was separated from the discarded supernatant (carefully pouring from the tube). The DNA precipitation procedures were repeated twice. The tubes were kept inverted (on paper towel) to dry completely. The air-dried pellet was resuspended in 50mL of TE buffer (10mM Tris, 1mM EDTA [pH 8]), carefully rubbing the pipette tip on the tube wall to release the DNA, which was kept at -20°C until use.
choE gene amplificationchoE gene amplification followed the protocol of Ladrón et al.13. A mixture of 2.5mL of 10 X PCR buffer; 0.75mL of 10mM deoxynucleoside triphosphate (DNTP); 0.75mL of 50mM MgCl2; 0.5mL of Taq DNA polymerase; 1mL (40pmol) of each primer; 10mL of DNA template; and 8.5mL milli Q water (final volume of 25mL) was used for PCR. In a thermal cycler (PTC-100, MJ Research), the DNA was first denatured at 95° C for five minutes and then subjected to 30 amplification cycles under the following conditions: denaturation at 95°C for one minute, annealing at 55° C for one minute, and extension at 72°C for one minute. After the final cycle, the reactions were terminated by an extra run at 72°C for 10minutes. Electrophoresis on horizontal 2% agarose gels was performed on 25mL of the amplified product and with 0.5mg of ethidium bromide per mL for two hours using TAE buffer (40mM Tris-acetate, 2 mMNa2 EDTA 2H2O) and an electric current of 100V. Fragments corresponding to the 959 pb product of choE gene amplification, indicative of R. equi, were photographed using AlphaImager (Alpha Innotech ®).
ResultsAmong the 78 bacterial isolates showing morphologic characteristics of Rhodococcus spp., 57 were presumptively identified as R. equi by a positive CAMP test reaction. However, the presence of the gene choE, indicated by the 959bp fragment amplification product, was observed in only 51 of these isolates, as well as in the reference strain R. equi ATCC 6939. Thus, six isolates CAMP test positive and 21 isolates CAMP test negative were also negative for the presence of the choE gene. Phenotypic characteristics assigned for R. equi were shown by all 51 isolates CAMP test-positive having a confirmed presence of the gene choE. Accordingly, these isolates did not utilize carbohydrates, acetate, citrate, or malonate as a sole carbon source, but produced catalase and lipase. In addition, all isolates showed negative results in tests for amylase, beta-galactosidase (ONPG), casease, DNase, gelatinase, esculinase, H2S, indole, oxidase, and motility. A variable behavior was demonstrated in the production of nitrate reductase, urease, and reduction of hippurate. While adenine was completely transformed, hypoxanthine, tyrosine, and xanthine were not. Other features consistent with R. equi included the optimal development of cultures in a strictly aerobic environment at temperatures between 25°C and 37°C. All these features were confirmed by the reference strain R. equi ATCC 693914,16.
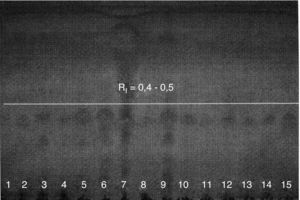
Fig. 1 shows representative results of mycolic-acid analysis by TLC. Spots corresponding to products having Rf around 0.4-0.5 were observed in all suspected isolates of R. equi and were comparable with the results from standard R. equi ATCC 6939. Other reference strains tested simultaneously for mycolic-acids yielded Rf values between 0.2-0.6, as follows: Corynebacterium pseudodiphthericum IAL 0104 and C. xerosis IAL 0105 - Rf 0.2; Nocardia asteroides IAL 2125, N. brasiliensis IAL 2126, N. brevicatena IAL 2123, Rhodococcus equi ATCC 33701, R. equi ATCC 33702, R. equi ATCC 33703 and R. equi ATCC 6939 - Rf between 0.4 and 0.5; Mycobacterium aurum ATCC 23366, M. vaccae ATCC 15483 and M. smegmatis ATCC14468 - Rf ≥ 0.6.
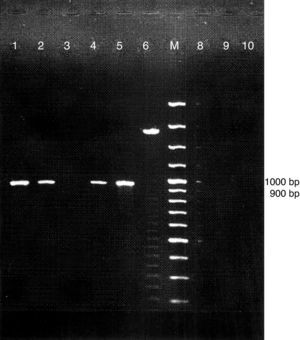
A representative eletrophoretic pattern of PCR amplification products is shown in Fig. 2. The presence of the expected choE gene 959bp fragment was observed in 51 isolates CAMP test positive showing phenotypic characteristics determined for R. equi, as well as in the reference strain R. equi ATCC 6939. In contrast, PCR results for 22 isolates CAMP test-negative, and five isolates CAMP test-positive were negative for the presence of the choE gene.
Representative eletroforetic pattern of PCR amplified products of choE in genomic DNA samples of suspected Rhodococcus equi. Lanes 3, 6, 8, 9 and 10 (isolates CAMP test negative); Lanes 1, 2 and 4 (isolates CAMP test positive); Lane 5 reference strain Rhodococcus equi ATCC 6939; M=DNA size marker (100bp).
R. equi is recognized as an opportunistic pathogen in humans and has emerged as an important cause of morbidity and mortality among immunocompromised patients, especially those infected with HIV. The increased awareness of rhodococcosis improves chances for rapid and correct diagnosis. Basic and clinical analysis is necessary for an appropriate treatment.
Seventy eight bacterial isolates suspected to be Rhodococcus spp. were investigated using phenotypic and genotypic tests, including TLC and PCR. A total of 51 isolates were confirmed as R. equi. The phenotypic identification was based on morphology, cultivation, nutrition, and biochemical features. The 51 isolates of R. equi had characteristics very similar to those described in the literature.4–6,8,20–22
All 51 isolates grew well when incubated in aerobic conditions at 37°C in a non-selective medium, MH agar. After incubation for 24hours, colonies measuring about one to two mm in diameter were not distinct. Colonies had their apparent features developed in 48hours with entire edges, shaped like irregular round teardrops, smooth, semitransparent, glistening, coalescent, and mucoid.
The characteristics described by Prescott6 for R. equi were investigated in the confirmed isolates. Colony variation was observed in cultures with the predominant classical type of coalescent viscous mucoid colonies, but less mucoid forms were also present. Pigment production was not observed in cultures less than four days old, but in up to seven days colonies developed a delicate salmon-pink color. Cultures maintained in slant medium for prolonged periods without sub-culture often became rough, dry, and orange-red, but reverted to the classic colony type when returned to sub-cultures. Morphologically, typical colonies of R. equi ATCC 6939 appeared smaller, smooth, dry rather than mucoid, and appeared to be more pigmented than the colonies of the suspected isolates of R. equi.
Regarding the microscopic morphology and staining properties, all suspect R. equi were Gram-positive pleomorphic coccobacillus, varying from distinctively coccoid to bacillary depending on growth conditions. Coccoid forms were usual on solid media (MH agar), but in liquid media (brain-heart infusion), particularly in young cultures, there were forms of long rods or short filaments, which sometimes showed rudimentary branching. When stained by the Ziehl-Neelsen Kinyoun-modified method, the isolates showed partial resistance to acid-alcohol, maintaining basically the same morphology observed in smears stained by the Gram method. All these characteristics are exactly as those related to R. equi by Prescott,6 and most isolates proved to be partially acid-fast. According to the author, this is an inconsistent feature of the microorganism.
Several features found in this study can be routinely used for identification of R. equi. All isolates produced the equi factor, emphasizing the importance of the CAMP test. This test was used as a phenotypic marker for the rapid presumptive identification of R. equi, but it may miss strains in isolates not expressing COX despite having the choE gene, or give false-positive results for other extra cellular COX-producing actinomycetes. As well as R. equi, other Gram-positive mycolic acid-containing bacteria, such as Mycobacterium spp., R. erythropolis, R. rhodochrous, and Dietzia spp. have COX23,24 activity and the possibility of presenting a positive result in the CAMP test. Nevertheless, the assay proved to be an important tool to presumptively identify R. equi, since the 51 isolates were CAMP test-positive. According to Prescott,6 the test is distinctive for this organism and must always be used in presumptive identification, since no R. equi isolate has been described as equi factor negative.
TLC, the method chosen to investigate mycolic-acids and recommended by Miyaji et al.,8 is the easiest to perform and is inexpensive. The usual revelation reagent was substituted in this study by 10% rhodamine in ethyl alcohol, which does not interfere with the detection of methyl-esters present. In the 51 isolates suspected to be R. equi and in the reference strain R. equi ATCC 6939, spots corresponding to mycolic acids were detected, with Rfs between 0.4 to 0.5, in agreement with the values obtained by Amat.7Other mycolic acid-containing bacteria belong to the genera Corynebacterium, Dietzia, Gordonia, Millisia, Nocardia, Segniliparus, Skermania, Smaragdicoccus, Tomitella, Tsukamurella, and Williamsia. Mycolic-acids are considered very important chemotaxonomic markers, and their identification by TLC is a useful tool, especially in the screening of members of some genera.25–30
According to Miyaji et al.,8 mycolic-acids based analysis allows categorization of some genera into three groups: group i- Mycobacterium and Tsukamurella, which present Rf values ≥ 0.6; group ii- Gordonia, Nocardia, and Rhodococcus, which present Rf between 0.2 - 0.6; and group iii- Corynebacterium, which presents Rf ≤ 0.2. According to these criteria, the results for mycolic-acids analysis in this study are compatible with R. equi identification and indicate that the TLC methodology may be standardized in this laboratory.
In the PCR assay used to detect the vapA gene for rapid molecular identification of R. equi,9,10,12,17 the sequences of 16S rDNA are generally accepted as a method for the differentiation of species, but heterogeneity may exist between different isolates of the same species.24,31 A study of the 16S rDNA sequences of several representative isolates has shown variations of above 4%, indicating that R. equi is a very heterogeneous taxon.32 Conversely, closely related species may have identical or nearly identical sequences of 16S rDNA.33,34 Therefore, other species-specific targets are needed, such as the choE gene, to undertake a reliable identification of the bacterial species. Taylor et al.34 developed a method of PCR-RFLP targeting a heat shock protein (hsp) gene of 65-kDa, which had previously been used for identification of mycobacteria, and was shown to be useful to discriminate isolates of R. equi. However, according to Ladrón et al.,13 the test is very laborious because of the conservative nature of this gene. The amplification product is of the same size for all species of actinomycetes, and R. equi can be discriminated by restriction analysis of amplicons.
The PCR method proposed by Ladrón et al.13 was chosen to confirm the identified R. equi in bacterial isolates that express the activity of COX through the CAMP Test. The results, indicated that sequences complementary to the primers COX were present in 51 isolates suspected of R. equi, suggesting that they are preserved in this bacterial species. In all 51 isolates, PCR results were consistent with the production of COX activity of the extracellular-derived choE gene that was expressed phenotypically by a positive reaction in the CAMP test. The choE gene was not detected in any of the 21 bacterial isolates that tested negative for the CAMP test. Moreover, among the 57 bacterial isolates that showed detectable activity in the CAMP test, six were negative for the choE gene, indicating that they probably belong to other species of actinomycetes.32 The data strengthen the validity of the method and the use of COX molecular primers to identify R. equi, especially when the CAMP test is positive. Thus, as suggested by Ladrón et al., the presence of the choE gene identified by a specific PCR method should be considered in the rapid and correct identification of R. equi, differentiating it from other pathogenic actinomycetes.
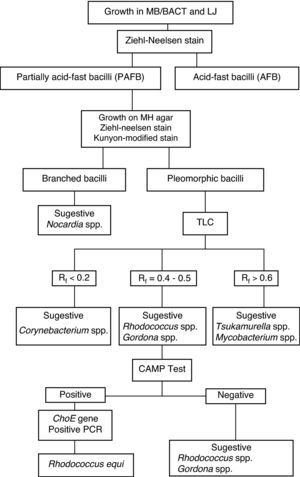
Results of the investigation of the 78 isolates from sputum suggest a simplified identification scheme for R. equi, as a first step in the search for partially acid-fast bacilli based on the determination of mycolic-acids by TLC technique, followed by the CAMP test and PCR using the COX primers that amplify the choE gene (Fig. 3).
According to the literature, R. equi is a frequent cause of cavitary infections in immunocompromised patients, but extra-pulmonary infections in immunocompetent patients have also occurred. Clinical symptoms may be insidious and resemble those of mycobacterial infections, fungi, and other actinomycetes. R. equi should always be suspected when cultures, especially those for tuberculosis, show colonies with distinct morphology and microscopic observations after Gram stain indicates the presence of Gram-positive pleomorphic coccobacilli. Staining of culture smears by the Ziehl-Neelsen Kinyoun-modified method and microscopic observations of partially acid-fast microorganisms strengthen a positive identification.
In many instances, identification of M. tuberculosis in routine clinical laboratory procedures can be faulty and lead the physician to start empirical treatment of tuberculosis, leaving a patient with untreated rhodococcosis. This study could be an alert to healthcare professionals about the importance of identifying R. equi, even presumptively, in human isolates.
The phenotypic identification was efficient to presumptively identify 57 bacterial isolates as R. equi, considering the colony and cellular morphology, biochemical tests, presence of compounds with Rf 0.4-0.5 by TLC, and especially the CAMP test reaction. The fastest methods were PCR and TLC, whereas the phenotypic characterization took five to 15 days. The differences are mainly due to the longer time required for bacterial growth and the large number of biochemical tests used. PCR was the more accurate method, correctly identifying 51 isolates of R. equi. The three methods used to analyze the reference strain of R. equi, ATCC 6939, produced similar results.
It is concluded that the identification of R. equi can be achieved by combining the three methods in an optimized algorithm (Fig. 3). Cultures and phenotypic identification including TLC should be considered as the gold standard method to detect the presence of this bacterial agent in the investigated clinical sample. Phenotypic tests and TLC should be performed on primary cultures to suggest presence of Rhodococcus spp., and implementation of the CAMP test should be used for presumptive R. equi identification. PCR should be considered an alternative or supplement method to provide highly specific and sensitive results that can be obtained faster. More rapid and accurate identification of the organism provides for correct treatment and faster recovery of the patient.
Conflict of interestAll authors declare to have no conflict of interest.