Staphylococcus aureus (SA) remains a major cause of nosocomial and community-acquired infections worldwide. Nasal carriage of this bacterium among hospital personnel constitutes an important source for nosocomial infections. A cross-sectional study enrolling the whole medical student population (n=387) of the School of Medicine at the Universidad de Cartagena, Colombia, was conducted to evaluate the carriage rates of both methicillin sensitive- and methicillin resistant-SA, the frequency of Panton-Valentine leukocidin genes in the isolates, and risk factors associated with carriage in this selected population. After signing an informed consent, participants completed a survey related to possible risk factors for colonization, and nasal swabs were collected from anterior nares. Staphylococcus aureus strains isolated from carriers were subjected to DNA extraction and PCR assays to determine the presence of MecA and Panton-Valentine leukocidin genes. Typing of the staphylococcal chromosomal cassette was performed for methicillin resistant strains. Molecular analysis was performed for only one strain per carrier. Prevalence of carriage for methicillin sensitive- and methicillin resistant-SA was 25% and 1.6% respectively. Most of the methicillin resistant isolates carried the staphylococcal chromosomal cassette type IV and the genes for Panton-Valentine leukocidin. To determine carrier types among medical students, each participant was subjected to four additional swabs, each taken two weeks apart. 9.8% persistent carriers, 53.1% intermittent carriers, and 37.1% non-carriers of SA were found. There was no association between risk factors analyzed and carriage of the bacterium. The study was conducted from April to September 2009 and found a persistent carriage of methicillin resistant-SA strains bearing the genes for Panton-Valentine leukocidin among medical students, evidencing the potential contribution of this portion of healthcare personnel either to the spread or introduction of these strains into the healthcare environment.
Staphylococcus aureus (SA) remains an important human pathogen responsible for health care- and community-associated infections, which also behaves as a commensal in healthy individuals, adopting the anterior nares as its ecological niche.1,2 A worldwide concern in public health is the emergence of methicillin-resistant SA (MRSA) strains, first considered to be confined to hospitals and medical centers and nowadays presenting in community settings. Cases of MRSA infections have been increasingly reported during the last decade in healthy individuals of the general population with no traditional risk factors for these community-associated MRSA (CA-MRSA) infections.3–5 CA-MRSA strains have been identified as highly clonal and virulent, responsible for over 30% of SA infections.6–8
It is well-known that nasal colonization constitutes a risk factor for subsequent severe infection, but it also can be the source of transmission of this bacterium to other susceptible individuals.1,9 Asymptomatic carriage of SA in healthy individuals has been shown to have a high prevalence, especially in children, young adults, and healthcare workers.10–12 Thus, colonized individuals in frequent contact with the general community or healthcare environment may spread the bacteria to other community members or to susceptible patients, respectively.13–16 Medical students represent an important portion of the healthcare personnel, and they are in frequent contact with patients; thus, they are at risk of being colonized with different pathogens, including SA, and of spreading them to susceptible patients.17,18 Different studies around the world17–21, in Colombia,22 and Brazil23 have evaluated nasal carriage rates of SA in medical students. Some of these studies reported that nasal carriage rates of SA increased with greater exposure of students to the hospital environment.17,20
Although still a matter of debate, both epidemiological and clinical studies point out that the high virulence potential of CA-MRSA originates from Panton-Valentine leukocidin (PVL), a two-component cytotoxin that targets human and rabbit polymorphonuclear neutrophils, and monocytes or macrophages, or both.3,24,25 Furthermore, recent studies indicate that the clinical sequelae of infections caused by PVL-positive SA strains, regardless of methicillin resistance, tend to be more severe than infections caused by PVL-negative strains.24,26,27
Although the role of PVL in CA-MRSA pathogenesis is still controversial, epidemiological and clinical data5,24 provide strong evidence that the high virulence potential of CA-MRSA is associated with the genes lukS-PV and lukF-PV encoding the subunits of PVL.
It has been shown that CA-MRSA isolates associated with disease carry the genes for PVL more frequently than CA-MRSA associated to asymptomatic colonization.28 Thus, carriers of CA-MRSA strains positive for PVL genes may pose an increased risk of spreading disease-causing strains to susceptible individuals. Furthermore, recent studies in different parts of the world have reported the asymptomatic carriage of MRSA strains positive for PVL genes in medical students, a fact that may contribute to the spread of MRSA between the community and hospitals.29 In contrast, in Colombia, there are no data about the frequency of PVL genes in nasal isolates in medical students or other health care workers. Thus, this study aimed to investigate nasal carriage of PVL-positive SA strains, carriage categories, and risk factors associated to colonization, in medical students from the Universidad de Cartagena, Colombia during 2009.
Material and methodsDesign and study populationThe study was performed from April to September, 2009 by the Genetics and Molecular Biology Research Group of the Universidad de Cartagena. This was a cross-sectional study enrolling the entire student population of the School of Medicine at the Universidad de Cartagena, Colombia, comprising 387 medical students, of which 372 completed the study. Students who agreed to participate were asked to sign an informed consent and complete a written questionnaire on demographics and medical history. Variables included in the questionnaire were age, gender, previous infections, allergies and other non-infectious pathologies, smoking habits, antibiotic usage in the previous three months, and surgeries and hospitalizations in the previous six months. The study was approved by the Ethics Review Board of the Universidad de Cartagena.
To determine SA carriage types in the study population, participants were subjected to four additional consecutive nasal swabs, each taken two weeks apart. Based on microbiological results, individuals were classified in three categories: non carriers, intermittent carriers, and persistent carriers, according to the definitions proposed by Kluytmas et al.9 Participants that yielded a total of five negative cultures for SA were classified as non-carriers; those yielding one to three positive cultures were classified as intermittent carriers; and those yielding four or five positive cultures were classified as persistent carriers.
Specimen collectionFor each participant, nasal swabs were taken from both nostrils, inoculated into Stuart's transport medium (OXOID, England), transported to the Laboratory of Microbiology of the School of Medicine at the Universidad de Cartagena, and processed within eight to 18hours according to previously described protocols.30 Briefly, specimens were plated onto mannitol-salt agar (BBL – Becton, USA) and incubated at 35°C–37°C for 48hours; mannitol-salt positive colonies were subcultured onto nutrient agar (Difco – Michigan, USA) at 37°C for 24–48hours. Identification of SA was based on colony morphology, gram stain, catalase test, tube coagulase test (BBL, coagulase plasma rabbit – Becton, USA), and latex agglutination test (Staphaurex – Remel, UK).
Antibiotic susceptibility testingAntibiotic susceptibility of SA strains was performed by the disc diffusion method following the recommendations of the Clinical and Laboratory Standards Institute (CLSI).31 The antibiotics evaluated were: rifampin, clindamycin, erythromycin, gentamicin, trimethoprim/sulfamethoxazole, cefoxitin, and oxacillin. The D-zone test for inducible clindamycin resistance was performed for each isolate according to the CLSI method.31 Isolates were classified as MRSA if they demonstrated resistance to cefoxitin, and as methicillin-susceptible SA (MSSA) if they were susceptible. For methicillin-resistant strains, susceptibility to vancomycin was further evaluated by the agar dilution method31 at concentrations of 0.5 to 32μg/mL, and MICs were measured manually after 24h of incubation. The SA strains ATCC-29213 (vancomycin-sensitive), ATCC-25923 (MSSA) and ATCC-33591 (MRSA) were used as control strains. MSSA and MRSA isolates were maintained frozen at -35°C for use in subsequent molecular assays.
Molecular analysisGenomic DNA extractionGenomic DNA from each isolate included in the study was obtained according to the protocol described by Millar et al.,32 with some modifications. Briefly, each SA isolate was streaked in nutrient agar and incubated for 24h at 37°C. Five colonies were then suspended in 1mL of Tris 0.5M, centrifuged at 13,000rpm x 5min. Supernatant was discarded and the pellet resuspended in 500μL buffer TE (10mM Tris; 1mM EDTA, pH:8.0) and boiled at 100°C for 30min, and then incubated at -35°C for 20minutes, thawed at 65°C, and finally centrifuged at 13,000rpm for 15min. Supernatant containing bacterial DNA was collected in a clean tube and stored at -20°C for subsequent PCR assays.
Detection of Nuc, MecA and PVL genesSA isolates identified by microbiological methods were subjected to multiplex polymerase chain reaction assays to assess the presence of lukF-PV (encoding part of the PVL toxin), mecA (methicillin resistance determinant), and nuc genes using a set of three primer pairs: MecA1F – MecA2R that amplifies a 147bp fragment of mecA gene;33 Nuc1F – Nuc2R that amplifies a 300bp fragment of nuc gene specific for SA,34 and LukPV1F – LukPV2R that amplifies a 437bp fragment of PVL gene.25 SA strains ATCC 33591 (mecA +; nuc +; PVL −) and ATCC 25923 (mecA −; nuc +; PVL +) were used as controls for mecA and PVL amplification and pure water was used as control for the PCR assay.
Typing of SCCmec elementAll confirmed MRSA isolates were subjected to SCCmec typing using a multiplex PCR assay in accordance with the protocol described by Zhang et al.33 using the SA control strains NCTC10442 (SCCmec type I), N315 (SCCmec type II), and JCSC4744 (SCCmec type IV).
For each PCR assay, DNA from each isolate was amplified in a reaction volume of 25μL containing 12.5μL of the PCR mix (PCR master mix; Promega®), 0.2uM of each primer, and 5μL of DNA template. The reaction was performed in a Perkin-Elmer thermal cycler under the following conditions: an initial denaturation step at 94°C for 5min, followed by 30 cycles of 94°C for 1min, 50°C for 1min, and 72°C for 2min, with a final extension at 72°C for 10min. PCR products were visualized in a 2% agarose gel stained with ethidium bromide under UV transillumination.
Statistical analysisStatistical analysis was performed with the Statistical Package for Social Sciences (SPSS) version 18.0 for Windows (SPSS Inc. – Chicago, IL). To determine the prevalence of SA nasal carriage, the study population was classified as carrier and non-carrier based on microbiology results obtained from the first nasal swab survey. Conversely, carrier types (non-carrier, persistent carrier, and intermittent carrier) were determined at the end of the study based on the definitions by Kluytmas et al.9 Univariate analysis was applied to determine the association of colonization or carrier types to potential risk factors using the chi-square test, with a p-value≤0.05 for statistical significance.
ResultsA total of 372 participants from the entire medical student population, comprised by a total of 387 students during the study period, were surveyed. The age of the participants ranged between 15 and 26 years (average 19 years, SD: ±2.21). At the first nasal swab survey, SA was isolated from 101 (27.15%) of the 372 participants; six (1.61%) participants were MRSA carriers, and 95 were MSSA carriers (25.5%). There was no statistical association with age (p=0.752) or gender (p=0.275). Furthermore, risk factors including contact with patients, previous hospitalizations, antibiotic usage, nasal abnormalities, allergic diseases (asthma, sinusitis, and rhinitis), smoking habits, and years of study at the university, were not associated to nasal carrier state of SA.
The 101 SA strains obtained at the first nasal swab survey were subjected to antibiotic susceptibility testing and molecular analysis. Six isolates were resistant to oxacillin (5.9%); three were resistant to clindamycin (2.97%), and 15 were resistant to erythromycin (14.85%), one of which had the inducible clindamycin resistance phenotype (D-zone test positive). No strain showed resistance to gentamicin, trimethoprim-sulfamethoxazole, or rifampin. None of the six MRSA strains were resistant to vancomycin (MIC<0.5μg/mL); one MRSA strain was erythromycin-resistant.
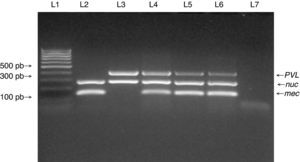
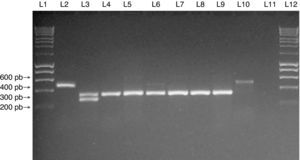
PVL genes were found in only four of the 95 MSSA strains, and in five of the six MRSA strains (Fig. 1). SCCmec typing of the MRSA strains showed that five carried the SCCmec type IV. The remaining MRSA strain was SCCmec type I (Fig. 2). The five MRSA strains bearing SCCmec type IV were also PVL-positive, while the strain bearing SCCmec type I was PVL negative. Thus, PVL genes were more frequently carried by MRSA strains than by MSSA strains (5/6 vs. 4/95, p≤0.0001).
Multiplex PCR for detection of nuc, mecA, and PVL genes. Multiplex PCR was performed to detect the presence of nuc, mecA and PVL genes in isolates from medical students to confirm Staphylococcus aureus species (nuc gene), methicillin resistance (mecA gene) and presence of PVL genes. Lane 1: MW (DNA molecular weight markers). Lane 2: ATCC 33591 reference strain (nuc+, mecA+, PVL-). Lane 3: ATCC 25923 reference strain (nuc+, mecA-, PVL+). Lanes 4 to 6: MRSA isolates from the study. Lane 7: negative control for the PCR reactions.
Multiplex PCR assay for SCCmec typing of SA isolates. Multiplex PCR was performed for typing of the SCCmec element in MRSA isolates from the study. Lanes 1 and 12: MW (DNA molecular weight markers). Lane 2: NCTC10442 reference strain (SCCmec type I). Lane 3: N315 reference strain (SCCmec type II). Lane 4: JCSC4744 reference strain (SCCmec type IV). Lanes 5 to 9: MRSA isolates from medical students carrying SCCmec type IV. Lane 10: MRSA isolate from a medical student carrying SCCmec type I. Lane 11: negative control for the PCR assay.
In addition, to determine SA carrier types in the study population, the 372 participants were subjected to four additional consecutive nasal swabs, each taken three weeks apart. From the 372 students subjected to five nasal swabs, 9.7% (n=36) were persistent carriers, 53.2% (n=198) were intermittent carriers, and 37.1% (n=138) were non-carriers. Three persistent carriers were colonized with MRSA strains that carried the genes for PVL and SCCmec type IV, and they represented medical students enrolled in semesters with clinical rotations.
DiscussionIn Colombia there are few reports about the prevalence of SA carriage in medical students,22 and reports on CA-MRSA colonization in individuals without risk factors associated to healthcare are scarce.30,35 Traditionally, colonization by MRSA has been evaluated and described in healthcare personnel, or individuals that have contact with these institutions or their workers (physicians, nurses, hospital workers, etc.). This study found that 1.61% of medical students from the Universidad de Cartagena carried in their nares, persistently or intermittently, MRSA strains with the community-associated profile (PVL-positive and SCCmec type IV), providing evidence that these strain types are circulating in the healthcare personnel in Cartagena. Previous studies in Colombia have shown MRSA colonization rates in the pharynx, nares, and on the hands similar to those reported here. For example, Ganoa et al.22 reported that 1.3% of medical students in clinical rotations carried MRSA strains in their nares, and Baez et al.35 recently reported that 0.6% of individuals from the general population with no apparent exposure to healthcare environment and no antibiotic usage in the previous six months carried CA-MRSA strains on their hands. Although the rates for MRSA carriage in healthcare personnel are relatively low, they are significant, since they provide evidence of colonization at different anatomical sites by CA-MRSA both in individuals from the general population and in medical students who are in close contact with patients during their clinical rotations in different hospital centers. This constitutes an important risk factor for spreading of the pathogen to patients and to the general population.
The present results are similar to those shown in international studies. For example, studies from United States have reported up to 1.53% of intermediate resistance to methicillin in SA isolates from medical students in first and third year.36 In Chile, Cifuentes et al.37 found that around 5.2% of medical students participating in their study were nasal carriers of MRSA. Baliga et al.,38 in Turkey, reported that 4.4% of medical students in clinical practice were MRSA carriers. In Brazil, Prates et al.23 reported that the prevalence of CA-MRSA nasal carriage in university students was 2.4%.
In Colombia, several studies have shown PVL-positive SA strains, both MSSA and MRSA, as important source of infections of varying severity,39–41 evidencing an increasingly serious public health problem in this country. The present data show that MRSA isolates from medical students share features of CA-MRSA (Figs. 1 and 2), warning about the potential risk for introduction of these strains to the healthcare environment.
Published evidence42,43 points out that health-care workers more likely act as vectors and not as the main sources of MRSA transmission; thus emphasizing that good hand hygiene practices remain essential to control the spread of MRSA. In addition, eradication of MRSA colonization from patients and healthy carriers has been tried with topical and oral antibiotics. For example, international guidelines recommend the use of mupirocin for nasal decolonization in certain groups of patients and health-care workers colonized with MRSA.44 However, this antibiotic is only effective at removing the bacterium from the nose over a few weeks, and nasal relapses are common within several months.45 Moreover, clinical studies have found little or no efficacy of mupirocin treatment in preventing nosocomial SA infections.46–48 Another shortcoming for this antibiotic is the increasing trend in resistance and the subsequent reduction in effectiveness.49,50
Since there are no recommended guidelines for routine decolonization of CA-MRSA in healthcare workers, especially medical students, and the authors were concerned about the reported rise in bacterial resistance to mupirocin, the most used antibiotic for nasal decolonization, it was decided not to recommend any sort of medical therapy to the group of CA-MRSA carriers in the student population. Instead, the three persistent CA-MRSA carriers identified in this study were notified about their colonization status and invited to participate in an educational session with emphasis on hand hygiene and standard precautions for proper handling of patients, aimed to decrease spread of the bacterium and to reduce the risk of infection in susceptible patients.42,51
ConclusionsCA-MRSA carriage was found in this medical student population, and PVL genes were significantly carried by these MRSA strains compared to MSSA strains (p≤0.0001). The different carrier types of SA colonization in medical students from this university were also identified. The high frequency of SA carriage (persistent and intermittent) found in this study may represent a risk for the spreading of SA strains, including MRSA strains, from the healthcare personnel to susceptible patients and to other individuals in the community.52 The present study demonstrates that medical students from this geographical area asymptomatically carry MRSA strains persistently, mainly PVL-positive, with the threat of spreading them both to community and hospital environments.
Studies addressed to determine the duration of CA-MRSA carriage, transmissibility, attack rates, and eradication strategies are required to clarify the role of health-care workers, including medical students, as reservoirs and vectors of these strains in health-care facilities.51 In addition, the reported increase in bacterial resistance to mupirocin, with its subsequent reduction in effectiveness, emphasizes the need for further studies aimed to develop strategies to successfully disrupt transmission of CA-MRSA strains to susceptible hosts.
Conflict of interestAll authors declare to have no conflict of interest.
This study was supported by the Universidad de Cartagena to Niradiz Reyes, PhD, Principal Investigator. The authors are thankful to the medical students that participated in the study; to Javier Escobar from the Laboratory of Bacterial Molecular Genetics at Universidad El Bosque, for technical assistance in the multiplex PCR assays; and to Juan Rebollo, for assistance in laboratory protocols.