Latent tuberculosis infection diagnosis based on the release of interferon-gamma in cultures of peripheral blood cells stimulated with Mycobacterium tuberculosis antigens has replaced the tuberculin skin test in many countries with low tuberculosis prevalence. The IFN-γ production can be influenced by genetic polymorphisms, of which the IFNG+874 (rs62559044) locus is the most studied. We investigated the possible influence of the IFNG+874 A/T polymorphism on interferon-gamma test performance.
MethodsPatients diagnosed with pulmonary tuberculosis (75), volunteers with positive tuberculin skin test (70) and healthy volunteers with negative tuberculin skin test and no history of contact with tuberculosis (57) were evaluated regarding the IFNG+874 genotype and the IFN-γ levels in whole blood cultures performed using an interferon-gamma commercial kit (QuantiFERON-TB® Gold In-Tube).
ResultsIFN-γ production was not influenced by the IFNG+874 genotype, regardless of antigen or mitogen-based stimulation, which suggests that other genes may influence IFN-γ production in response to mycobacteria. The IFNG+874 polymorphism was found to exert no influence over QFT-IT test sensitivity in our study.
ConclusionsThe IFNG+874 polymorphism was not shown to influence QuantiFERON-TB® Gold In-Tube test performance in an admixed population from northeastern Brazil.
Mycobacterium tuberculosis (Mtb) infection is present in one-third of the world's population. While some of these individuals are in a subclinical stage of tuberculosis (TB), most infected people will remain asymptomatic throughout their lives.1 These individuals are considered to have latent tuberculosis (LTBI) and only a fraction will develop active TB. This latent pool complicates disease control, as reactivation can occur at any time and chemoprophylaxis is administered only in individuals of known recent contact with an index case.2,3 LTBI screening is performed using tests based on an immune response to mycobacterial antigens. The tuberculin skin test (TST) has been used for over a century and offers good sensitivity and specificity in endemic conditions. However, it may present cross-reactivity to sensitization with the BCG vaccine as well as environmental mycobacteria.4,5 IFN-γ release assays (IGRAs) were first described in 20006,7 and present an alternative to TST.8 To date, most studies interpret IGRAs similarly to TST, except in populations vaccinated with BCG after childhood, or in individuals receiving multiple doses of this vaccine, as well as in locations where environmental mycobacteria infections are common.4,5 In these situations, IGRAs do seem to present a better positive predictive value due to greater specificity.9,10 Cost-effectiveness is another criterion to select IGRA as the test of choice, e.g. in countries with low TB prevalence.11–13
Both TST and IGRA tests are not able to distinguish active from latent infection. This is partly because these tests are based on the immune response, and those at increased risk to develop active TB are immunocompromised individuals who also present the highest rates of false-negative results. Moreover, the antigens expressed by Mtb during latent infection may not be the same as those expressed during the active replication stage.2,14 As a result, a lower specificity of TB diagnosis is expected in countries with high TB burdens.15–17
Studies focusing on the impact of genetic background with regard to IGRA testing are surprisingly scarce. IFN-γ production may be influenced by the presence of polymorphisms in the gene that encodes this cytokine, of which the most studied is the IFNG+874 A/T polymorphism. The IFNG+874 A/T polymorphism determines the emergence of an NF-κB binding site that can increase the transcription of the human IFN-γ gene.18 The AA genotype at this locus has been linked to TB susceptibility and low IFN-γ production, including in the Brazilian population.19 In a study among South African families in a hyperendemic area for tuberculosis, the influence of heredity on quantitative responses to IFN-γ release was estimated to be between 43% and 58%, depending on the nature of the antigen used as a stimulus20 It is possible, therefore, that the IFNG+874 A/T polymorphism influences the sensitivity and specificity values of IGRA assays. In this context, the aim of this study was to evaluate the influence of the IFNG+874 A/T polymorphism in M. tuberculosis infected individuals residing in an endemic area.
MethodsRecruitment and study designThe present study included 202 volunteers, divided into three groups: (i) patients with a diagnosis of pulmonary tuberculosis by positive culture and/or BAAR staining who were naïve to antituberculosis treatment (TB, n=75); (ii) volunteers with a positive TST greater than 10mm, with known contact with TB patients, who tested negative under culture and BAAR staining (TST+, n=70); (iii) volunteers with a negative TST, an absence of tuberculosis symptoms and no history of contact with individuals with tuberculosis (TST−, n=57). All individuals were recruited from two local reference institutions for TB diagnosis located in the city of Salvador, Bahia-Brazil. Written informed consent was obtained from all participants. This study was approved by the Institutional Review Board of the Bahiana School of Medicine and Public Health (registered under protocol: CAAE 57662016.8.1001.5662) and complied with the Declaration of Helsinki, as well as with Brazilian regulations pertaining to research ethics involving human beings (Resolution: CNS 466/2012).
Latent TB infection diagnosisLatent TB infection was diagnosed by TST (PPD RT 23 2TU, Statens Serum Institute, Denmark) and IGRA using a QuantiFERON®-TB Gold in tube (QFT-IT) (Cellestis Ltd., VIC, Australia) assay in accordance with manufacturers’ recommendations. Individuals were considered positive when presenting an increment of at least 0.35IU/mL in IFN-γ production in the Ag tube (specific antigen-stimulated culture) as compared to the Nil tube (cultures in the absence of any antigen), and an increment in IFN-γ production of at least 0.5IU/mL when comparing the PHA tube (mitogen-stimulated culture) with the Nil tube. Individuals were considered positive by TST when presenting skin induration measuring at least 10mm. QFT-IT was repeated for samples with discordant TST and IGRA results.
Genotyping and IFN-γ productionFollowing whole blood sample collection, genomic DNA was extracted using a PureLink® Genomic DNA Kit (Invitrogen, Carlsbad, CA, USA) in accordance with manufacturer's instructions. Genotyping for the IFN-γ polymorphism was performed using a Cytokine Genotyping Tray (One Lambda, Inc., Canoga Park, CA-USA) according to the manufacturer's protocols. The Human Th1/Th211plex FlowCytomix Multiplex Kit (eBioscience, Vienna, Austria) was employed in accordance with manufacturer's instructions to measure IFN-γ in pg/mL.
To correlate genotype and IFN-γ production in the supernatant of samples, we analyzed both the total IFN-γ production in each culture condition (pg/mL) and the differences between the IFN-γ levels (ΔIU/mL) in the Ag tube and the Nil tube of the volunteers enrolled, stratified by genotype.
Statistical analysesVariables were described as frequencies, percentages and median [interquartile range, in square brackets] values. The Chi-square test was used to evaluate the study population for the Hardy-Weinberg equilibrium, and to compare genotype and allele frequencies among groups. The Mann–Whitney test was used to compare IFN-γ production levels among different IFNG+874 A/T genotypes. The Pearson Chi-square test was used to compare proportions of each genotype among individuals with positive versus negative QFT-IT. Statistical significance was considered with an alpha error of 5%. Kappa index, odds ratio (OR) and sensitivity were calculated, with a confidence interval (CI) of 95%. Statistical analyses were performed using Prism software version 5 (GraphPad Inc., San Diego, CA) and R Project version 2.15.2 (GNU Project, Boston, MA).
ResultsClinical characteristics and frequency distribution of IFNG+874 A/T polymorphism genotypesThe frequency distribution of the IFNG+874 A/T polymorphism genotypes was in Hardy–Weinberg equilibrium for all groups (p=0.78). The IFNG+874 AA genotype was more frequently found among individuals with TB (chi-squared=10.94, p=0.004) when compared with TST− individuals (Table 1). The frequency of the A allele was also higher in TB patients than in TST- volunteers (chi-squared=17.6; p=0.001). Age and BCG scar positivity were similar among the different groups studied and among the IFNG+874 A/T polymorphism genotypes.
Frequency distribution of genotypes and alleles of IFNG+874 A/T polymorphism according to age, sex and ethnic background.
| TB | TST+ | TST− | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA (n=46) | TA/TT (n=30) | A (n=118) | T (n=34) | AA (n=34) | TA/TT (n=36) | A (n=97) | T (n=43) | AA (n=17) | TA/TT (n=41) | A (n=65) | T (n=51) | |
| Male (%) | 26 (66.7) | 13 (33.3) | 63 (80.8) | 15 (19.2) | 17 (60.7) | 11 (39.3) | 42 (75.0) | 14 (25.0) | 8 (34.8) | 15 (65.2) | 26 (56.5) | 20 (43.5) |
| Female (%) | 20 (54.1) | 17 (45.9) | 55 (74.3) | 19 (25.7) | 17 (40.5) | 25 (59.5) | 55 (65.5) | 29 (34.5) | 9 (25.7) | 26 (74.3) | 39 (55.7) | 31 (44.3) |
| White (%) | 4 (100.0) | 0 (0.0) | 8 (100.0) | 0 (0.0) | 3 (37.5) | 5 (62.5) | 10 (62.5) | 6 (37.5) | 4 (-26.7) | 11 (73.3) | 16 (53.3) | 14 (46.7) |
| Non-White (%) | 26 (54.2) | 22 (45.8) | 71 (74.0) | 25 (26.0) | 31 (50.0) | 31 (50.0) | 87 (70.2) | 37 (29.8) | 13 (30.2) | 30 (69.8) | 49 (57.0) | 37 (43.0) |
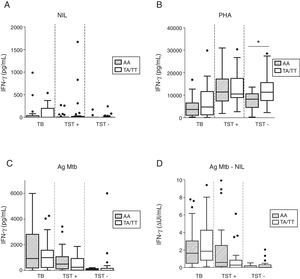
No significant differences were observed with respect to unstimulated IFN-γ production among the different groups and genotypes studied (Fig. 1A). IFN-γ production in response to polyclonal phytohemagglutinin (PHA) stimulation was lower among patients with pulmonary TB (3848 [1143–9120] pg/ml) in comparison to TST+ (11,080 [7462–17,248] pg/ml, p<0.0001) and TST− individuals (10,852 [6782–13,684] pg/ml, p<0.0001). Healthy uninfected individuals with the IFNG+874 AA genotype exhibited lower IFN-γ production upon stimulation with PHA when compared to IFNG+874 TT/TA genotype individuals (p=0.012, Fig. 1B). This difference was not significant when the IFN-γ levels were normalized by the total CD3+ cell counts (median values of 4.22 [2.84–9.12] for AA genotype individuals versus 6.24 [4.08–7.95] for those with TT/TA genotypes, p=0.3866), but these data were available for only 33 individuals of this group. Irrespective of genotype, no significant differences were seen in IFN-γ production under stimulation with PHA among the infected individuals (Fig. 1B). IFN-γ production upon mycobacterial antigen stimulation was not affected by genetic background at the IFNG+874 locus (Fig. 1C). Similarly, no significant differences were detected when analyzing QFT-IT test results (ΔUI/mL), i.e. mycobacterial antigen-stimulated versus unstimulated IFN-γ production (Fig. 1D).
IFN-production by pulmonary TB, TST+ and TST− individuals according to IFNG+874 genotype. (A) Non-stimulated IFN-γ production (NIL); (B) PHA-stimulated IFN-γ production as measured in the supernatant (PHA); (C) Mycobacterial antigen-stimulated IFN-γ production (Ag); (D) Difference in units (ΔUI/mL) between mycobacterial antigen-stimulated and unstimulated IFN-γ production.
No significant differences were observed with respect to the proportions of the different IFNG+874 A/T polymorphism genotypes when comparing QFT-IT-positive and QFT-IT-negative individuals (p=0.20, p=0.50 and p=0.28 for TST−, TST+ and TB, respectively, Table 2).
QFT-IT test results according to IFNG+874 A/T genotype.
| AA | TA+TT | Total | p-Value | |
|---|---|---|---|---|
| TB | ||||
| QFT-IT positive (%) | 43 (63.2) | 25 (36.8) | 68 | 0.4192 |
| QFT-IT negative (%) | 3 (42.9) | 4 (57.1) | 7 | |
| TST+ | ||||
| QFT-IT positive (%) | 22 (52.4) | 20 (47.6) | 42 | 0.0729 |
| QFT-IT negative (%) | 12 (31.6) | 26 (68.4) | 38 | |
| TST− | ||||
| QFT-IT positive (%) | 3 (27.2) | 8 (72.7) | 11 | 1.0000 |
| QFT-IT negative (%) | 14 (30.4) | 32 (69.6) | 46 | |
QFT-IT was found to detect TB with sensitivity of 90.7% (81.97–95.41) and specificity of 80.7% (68.66–88.87). In the TB group, the QFT-IT test presented similar sensitivity in patients of the AA genotype of the IFNG+874 polymorphism (93.5%, 82.5–97.8%) in comparison to those of the TT/TA genotype (86.2%, 69.5–94.5%). The odds of detecting TB were similar for AA genotype and TT/TA genotype individuals (OR=2.27, 0.435–13.03). Among AA genotype individuals, 3/46 TB patients presented false-negative QFT-IT results, while 4/29 individuals of either TT or TA genotype had false-negative QFT-IT results.
TST and QFT-IT presented regular agreement (kappa=0.34), with similar results when stratified by IFNG+874 A/T polymorphism genotypes (0.33 for the AA genotype and 0.28 for the TT/TA genotype).
DiscussionThe QFT-IT diagnostic values obtained in our study were lower than previously reported in other studies,10,21 yet consistent with previous findings in Brazil,22 as well as in other countries.23–25 QFT-IT sensitivity and specificity to detect TB was similar to values previously reported in countries with moderate disease burden.8,11,26
As previously reported, the frequency of the AA genotype and the A allele was higher in TB patients than in TST− volunteers.19
In the present study, only the group of healthy individuals presented differences in IFN-γ production following PHA stimulation according to genotype, with the lowest production found in IFNG+874 AA genotype individuals, as expected.18 This difference was not significant when IFN-γ levels were normalized by number of CD3+ lymphocytes. By contrast, no differences in IFN-γ production were found among the different genotypes within the infected groups evaluated, regardless of stimulation with either Mtb or PHA, which is consistent with previously reported findings,27 yet stands in contrast to other reports indicating that AA genotype TB patients had lower IFN-γ production in mycobacterial antigen-stimulated cultures.28–30 This seems to suggest that, both in the population studied, as well as in mycobacterial antigen-stimulated cultures, the IFNG+874 locus had no influence over IFN-γ levels under mycobacterial stimulation (in vivo or in vitro), as similar levels were detected regardless of genotype. The estimated heritability of the IFN-γ response to ESAT-6 was 58%, whereas the estimated heritability for BCG was 43%.19,31 It has been suggested that other genetic factors may influence anti-mycobacterial IFN-γ production in Mtb infection/disease.32,33 Despite the fact that two loci have recently been associated with IFN-γ production in response to ESAT-6 and BCG, the IFNG gene is not present in these regions.34
Similar differences in IFN-γ production were seen between the antigen-stimulated and unstimulated wells when comparing AA and IFNG+874 TT/TA genotype individuals. In Turkey, a study focused on the impact of IFNG+874 polymorphism on IFN-γ production in TB patients with varied clinical presentations showed a lower percentage of patients with detectable IFN-γ production among those of the AA genotype. This stands in contrast to our findings, as the proportion of responsive AA genotype individuals was similar to that of other genotypes evaluated.27
A recent Canadian study investigated possible influences of the IFNG+874 polymorphism on QFT-IT results. Using these authors’ published results, we calculated TB patient QFT-IT test sensitivity at 80% (64.1–90.0) among AA genotype individuals, compared to 75% (53.1–88.8) for TT/TA genotype individuals.35 However, when combining these authors’ previously published results with the present findings, the number of individuals evaluated was insufficient to demonstrate a relevant difference in sensitivity when comparing among the IFNG+874 genotypes.
ConclusionsIFNG+874 polymorphism had no influence over QFT-IT test performance in an admixed population from northeastern Brazil. Additional study is required to further elucidate any possible effect on QFT-IT test results arising from other genetic variations.
Conflicts of interestThe authors declare no conflicts of interest.
Financial supportConselho Nacional de Desenvolvimento Científico e Tecnológico (INCT-DT/MCT/CNPq 573839/2008-5) and Fundação de Amparo à Pesquisa do Estado da Bahia (PET035/2013).
To Yasmin S. de Azevedo Oliveira, Ana Paula Torres, Alice S. Martins dos Santos and Cleidiane Borges Daltro in contributing for technical support. To the team of the Laboratory of Immunology and Molecular Biology (Institute of Health Sciences/UFBA) for technical assistance. To the scientific coordination of Instituto Brasileiro para a Investigação da Tuberculose for their help with the logistics regarding the field work. To Andris Walter for expert language review of the manuscript.