The current epidemic proportions of infections caused by Staphylococcus aureus strains and especially by methicillin-resistant S. aureus (MRSA) are one of today's many threats to global public health, particularly in underdeveloped countries where significant gaps on the subject exist. The rapid spread and diversification of pandemic clones that exhibit remarkably increasing virulence and antimicrobial resistance pose a risk to the effective prevention and treatment of a wide range of infections. Undoubtedly, the remarkable versatility involving the pathogenesis and resistance of these bacteria is perpetuated through geographic and temporal factors inherent to clonal evolution and is reflected in the dramatic epidemiological changes of MRSA which, after decades prevailing in healthcare settings, have emerged in the community. Denominated community-associated [CA]-MRSA, these strains are particularly prevalent in some population groups, facilitating the spread of successful clones that are potentially capable of triggering severe community-acquired infections. Therefore, a broad approach to local epidemiological aspects in less studied regions, but nonetheless at latent risk of endemic spread that may reach global proportions, is necessary. In Brazil, despite limited molecular epidemiology data, CA-MRSA strains predominantly characterized as SCCmec IV, often classified as CC30-ST30, CC5-ST5 and CC8-ST8, seem to be spreading across different population groups in different regions of the country. Another important fact addressed in this review is the identification of the ST398-MRSA-IV/V clone and methicillin-susceptible S. aureus (MSSA) in healthy individuals from the community. Although susceptible to methicillin, the ST398 clone is associated with severe infections in humans and animals, denominated livestock-associated MRSA. It is therefore important to encourage assertive actions by all government sectors and by society, with a reassessment of current public health measures in light of the new perspectives arising from the scientific and epidemiological data on MRSA.
The epidemiology of Staphylococcus aureus has undergone a conceptual revolution in recent decades. This phenomenon was due in part to important changes in the epidemiological behavior of these bacteria, but also to a reassessment of old concepts in view of new knowledge arising from clinical and experimental research. Two key factors support the contemporary perspective of S. aureus, virulence and antimicrobial resistance. Therefore, a broad and simultaneous approach to these phenomena and the evaluation of their impact on the population are necessary, especially in developing countries such as Brazil where available data are often limited.
Severe infections caused by bacteria that are resistant to the antibiotics commonly used in clinical practice have emerged as a global health problem in the 21st century. Nowadays, antimicrobial resistance is a complex and multifactorial problem that affects society as a whole. Population characteristics, living conditions, agglomerations, injectable drug use, misuse of antibiotics, and underlying diseases all contribute to the selection and perpetuation of resistant bacteria. The current scenario of antimicrobial resistance is a matter of concern and tends to progress to what the World Health Organization calls the “post-antibiotic era”, which may be the near future.1
We continue to fail to contain the spread of resistance genes and we now have clinically important and potentially problematic organisms circulating in the community. Particularly methicillin-resistant S. aureus (MRSA) represent one of today's many threats to global public health because of the rapid spread and diversification of pandemic clones that exhibit remarkably increasing virulence and antimicrobial resistance.
Emergence of community-associated Staphylococcus aureus and virulence and resistance determinantsThe isolation of MRSA was described for the first time in England in 1961.2 In subsequent decades, the prevalence and epidemiology of MRSA have undergone dramatic changes, particularly in the 1990s, with an increase in the number of reports of infections associated with genetically distinct strains originating in the community.3 Thus, MRSA, which until then exclusive agents of healthcare-associated infections (HAIs), started to be recognized as the causal agents of severe disease acquired in the community and were later denominated community-associated [CA]-MRSA.4
For a long time, the clinical definition of CA-MRSA infections has been based on the absence of risk factors for hospital-associated MRSA (HA-MRSA) infection, such as a history of recent hospitalization and medical procedures such as dialysis, surgery or catheter use, and many studies continue to use these criteria.5 Furthermore, different genetic markers are strongly associated with strains typically defined as HA-MRSA or CA-MRSA. These markers will be addressed below.
The genetic determinant of methicillin resistance in MRSA, the mecA gene (which encodes a penicillin binding protein with low affinity, PBP2a), is transported by a mobile genetic element called staphylococcal cassette chromosome mec (SCCmec) which, by inference, confers resistance to all other β-lactam antibiotics, except for ceftaroline and ceftobiprole. The latter are fifth-generation cephalosporins indicated for the treatment of infections caused by MRSA.6 The acquisition via horizontal gene transfer and insertion of SCCmec into the chromosome of susceptible strains lead to the emergence of resistant staphylococcal strains. The high diversity in the structural organization and genetic content of these elements allows to trace the evolutionary origin of MRSA clones.7,8
Characteristically, the largest SCCmec types (I, II and III) are associated with HA-MRSA isolates, while SCCmec types IV and V, which are smaller, are frequently carried by typical isolates of CA-MRSA.9,10 However, with the rising prevalence and global spread of MRSA in the community over the past three decades, the epidemiological and molecular distinction of the origin of these strains has become blurred, with both CA-MRSA and HA-MRSA strains circulating in the community and concomitantly acting as nosocomial pathogens.7,11
The virulence and pathogenesis of CA-MRSA is somewhat complex and several factors probably contribute to the fitness of these strains during colonization and spread in the population. Studies suggest that the increased virulence of these strains may be associated with their capacity to evade death by neutrophils, the increased production of phenol-soluble modulins, high activity of the accessory gene regulator (Agr) system, the presence of SCCmec of low fitness cost, and a specific repertoire of toxins.4,7 In addition, the production of Panton-Valentine leucocidin (PVL), a cytotoxin that causes tissue necrosis and leukocytes destruction, was originally considered an important determinant of CA-MRSA virulence, playing a key role in the pathogenesis of necrotizing pneumonia.7,8,12 However, it is believed that its importance has been overestimated since PVL exerts its aggravating potential only in specific types and scenarios of infection, with the prevalence varying between strains and geographic areas, a fact suggesting that other virulence factors contribute to this process.4,7,12
Molecular epidemiology of CA-MRSA in BrazilSince their emergence and isolation, CA-MRSA strains have evolved differently in different geographic areas, a fact that has rendered the global epidemiology of these pathogens remarkably heterogeneous, with successful clones predominating in certain regions. This versatility is not only geographical but also temporal, with epidemic clones being replaced with emerging lineages following a pattern of clonal evolution over time.7
Certainly, the current situation related to these resistant strains has taken notorious proportions that require the development and improvement of tools capable of distinguishing isolated strains and of outlining the epidemiological panorama of S. aureus in the world and over time. In addition to SCCmec typing, some methods are useful to identify and to screen for different MRSA clones, particularly multilocus sequence typing (MLST). This method permits to infer clonal relationships between S. aureus isolates described in different parts of the world based on allele profiles classified as sequence types (ST), which can be grouped into clusters, the so-called clonal complexes (CC).
In Brazil, the first descriptions of CA-MRSA infections date back to the early 2000s and were reported in outpatients without a history of hospitalization or surgery from Porto Alegre, southern region of the country. These isolates belonged to the Oceania Southwest Pacific (OSPC) clone, ST-30-MRSA- IV.13 The same clone was isolated in Rio de Janeiro between 2004 and 2006, together with the USA400 clone (ST-1-MRSA-IV); the latter is associated with HAIs.14 The pandemic clone USA300 (ST-8-MRSA-IV), which caused respiratory infection in a patient from Porto Alegre, was also detected in the same study.14 Both studies identified isolates carrying the lukF gene, which encodes PVL.
Until the rise of CA-MRSA, the multidrug-resistant clone ST239-SCCmecIIIA, known as the Brazilian epidemic clone (BEC), and its variants predominated as nosocomial pathogens throughout the country.15–17 In the early 2000s, the ST5-SCCmecIV clone, known as the pediatric clone, which was not multidrug-resistant at first, was identified in Brazilian hospitals.18,19 Subsequently, this lineage seems to have developed multidrug resistance, as well as important virulence factors such as biofilm formation and enterotoxin production.15,20
Nowadays, in Latin America, SCCmec type IV is frequently detected in clones circulating in the community and in healthcare settings.21 In Brazil, infections with CA-MRSA have been reported in the cities of Porto Alegre, Rio de Janeiro, São Paulo, Botucatu, Recife, and Salvador, which ranged from skin and soft tissue infections to necrotizing pneumonia and severe sepsis. These infections were associated with clones harboring SCCmec type IV, similar to the OSPC clone (ST-30-MRSA-IV), and frequently carrying PVL-coding genes.13,14,22–28
Our research group also described a case of systemic CA-MRSA infection in an adolescent living in a small town in the interior of São Paulo (Bofete/SP), who had no history of healthcare exposure or recent travel.27 The isolate was characterized as ST5-MRSA-IV, carrying the genes encoding PVL and enterotoxin, and belonged to the same clonal complex as a lineage described in Argentina29 that is genetically related to the pediatric clone. These findings alert to the presence of CA-MRSA in small towns in rural Brazil and reinforce the need for comprehensive studies that contribute to elucidating the prevalence of these clones. Certainly, the spread of successful clones, especially in reservoirs of the community, facilitates the transport of endemic strains into households and puts the population at increased risk of infection.30
Community prevalence of Staphylococcus aureus and CA-MRSAInfections with CA-MRSA are particularly prevalent in some population groups. These groups include sportsmen, men who have sex with men, prison inmates, injection drug users, military personnel, children, people in correctional facilities or shelters, people living in overcrowded or low socioeconomic conditions, HIV-infected patients, and members of remote populations (such as Australian Pygmies and Aborigines), as well as American Indians, Alaskan Natives, and Pacific Islanders.31 However, it should be noted that all of these data are derived from sparse studies conducted in different countries. The studies are diverse in terms of objectives and methods and do not allow to draw a complete and coherent picture of the global epidemiology of CA-MRSA. In addition, there are significant gaps, with limited information for regions in Africa and South America, and the imminent endemic spread of CA- MRSA in underdeveloped countries has raised concern regarding probable devastating global consequences.7
Studies on the prevalence of asymptomatic S. aureus carriage are important since colonization is a possible precursor stage of invasive disease.32 Furthermore, a model developed by Macal et al.33 to study the dynamics of transmission of and infection with CA-MRSA in Chicago demonstrated that the majority of transmission events of these strains originated from asymptomatic carriers of MRSA and, less frequently, from individuals with active infection. This fact is interesting, considering that colonization is more common and usually lasts longer compared to infection, and should encourage the reassessment of public measures focused only on the control of clinically apparent infections.33
In 2011, our research group conducted a population-based survey that identified a prevalence of asymptomatic nasal S. aureus carriers of 32.7%, with six out of 686 urban residents of Botucatu, São Paulo, Brazil, being colonized with CA-MRSA (prevalence of 0.9%).34 All isolates carried SCCmec type IV and belonged to CC5. The study also estimated more than 1,000 MRSA carriers in the city.
Our findings agree with the results of the National Health and Nutrition Examination Survey (NHANES) which, in 2001-2002, estimated a 30.8% prevalence of S. aureus in the community and a 0.8% prevalence of colonization with MRSA in the United States.35 In subsequent years, more precisely in 2003-2004, the estimated prevalence of S. aureus exhibited a statistically significant reduction, dropping to 27.1%. On the other hand, MRSA colonization rates almost doubled during the same period (from 0.8% to 1.5%).36 It should be noted that, up to 2001, the most prevalent CA- MRSA clone in the United States was USA400 (ST1-SCCmec IV), which was largely replaced by the USA300 clone (ST8-SCCmec IVa), one of the most successful clones of all time,8 although it is currently declining.37
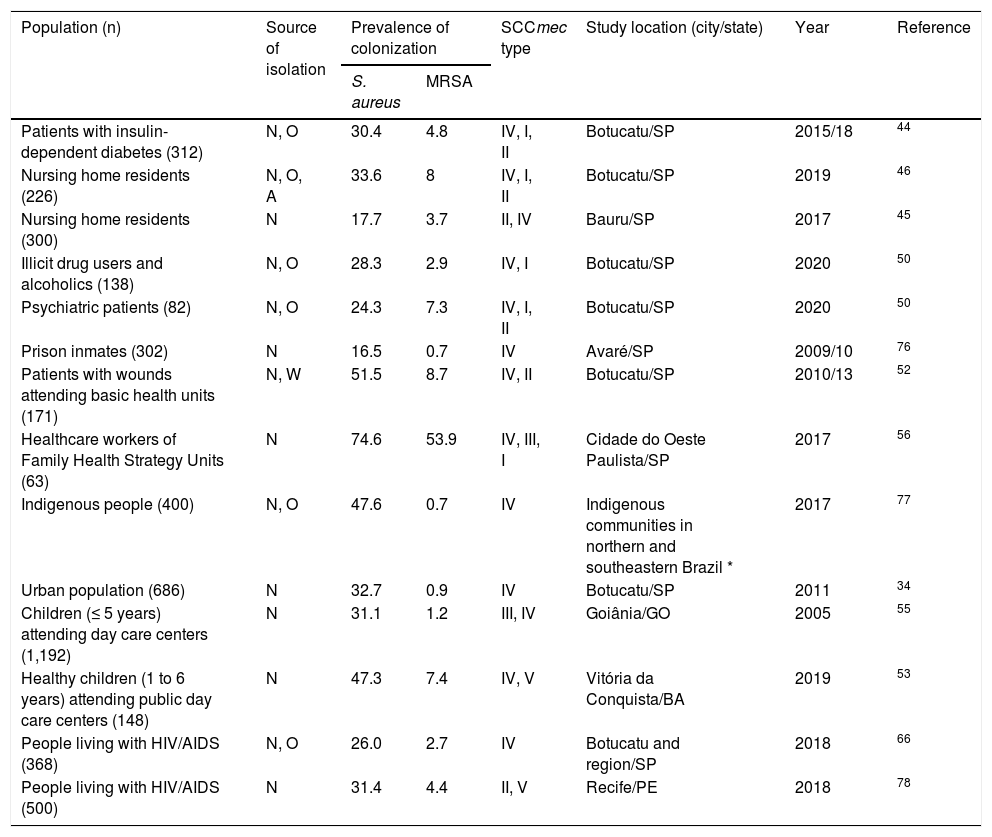
Other studies were conducted in Brazil to investigate asymptomatic MRSA colonization in different population groups. Some of those studies provided data on clonal characterization, which are summarized in Table 1. The majority of studies are concentrated in the state of São Paulo and generally indicate a rate of S. aureus colonization very similar to that described in the population-based study conducted in Botucatu, with the observation of higher rates among users of primary healthcare services, children attending public day care centers, and indigenous people. On the other hand, despite the correlated predominance of SCCmec IV, the prevalence of MRSA carriage was higher in the other studies, except for one involving prison inmates and indigenous people. These results are somewhat intriguing and disagree with data previously described for prisoners, American Indians and Australian Aborigines.36,38
Prevalence of MRSA colonization outside hospital settings reported in Brazil.
| Population (n) | Source of isolation | Prevalence of colonization | SCCmec type | Study location (city/state) | Year | Reference | |
|---|---|---|---|---|---|---|---|
| S. aureus | MRSA | ||||||
| Patients with insulin- dependent diabetes (312) | N, O | 30.4 | 4.8 | IV, I, II | Botucatu/SP | 2015/18 | 44 |
| Nursing home residents (226) | N, O, A | 33.6 | 8 | IV, I, II | Botucatu/SP | 2019 | 46 |
| Nursing home residents (300) | N | 17.7 | 3.7 | II, IV | Bauru/SP | 2017 | 45 |
| Illicit drug users and alcoholics (138) | N, O | 28.3 | 2.9 | IV, I | Botucatu/SP | 2020 | 50 |
| Psychiatric patients (82) | N, O | 24.3 | 7.3 | IV, I, II | Botucatu/SP | 2020 | 50 |
| Prison inmates (302) | N | 16.5 | 0.7 | IV | Avaré/SP | 2009/10 | 76 |
| Patients with wounds attending basic health units (171) | N, W | 51.5 | 8.7 | IV, II | Botucatu/SP | 2010/13 | 52 |
| Healthcare workers of Family Health Strategy Units (63) | N | 74.6 | 53.9 | IV, III, I | Cidade do Oeste Paulista/SP | 2017 | 56 |
| Indigenous people (400) | N, O | 47.6 | 0.7 | IV | Indigenous communities in northern and southeastern Brazil * | 2017 | 77 |
| Urban population (686) | N | 32.7 | 0.9 | IV | Botucatu/SP | 2011 | 34 |
| Children (≤ 5 years) attending day care centers (1,192) | N | 31.1 | 1.2 | III, IV | Goiânia/GO | 2005 | 55 |
| Healthy children (1 to 6 years) attending public day care centers (148) | N | 47.3 | 7.4 | IV, V | Vitória da Conquista/BA | 2019 | 53 |
| People living with HIV/AIDS (368) | N, O | 26.0 | 2.7 | IV | Botucatu and region/SP | 2018 | 66 |
| People living with HIV/AIDS (500) | N | 31.4 | 4.4 | II, V | Recife/PE | 2018 | 78 |
N: nasal; O: oropharynx; A: anal; W: wounds; n: number of subjects; SP: São Paulo (southeastern region); GO: Goiás (midwest region); BA: Bahia (northeastern region).
An important facet of the epidemiology of S. aureus is the fact that these bacteria infect “special populations”. This term refers to population strata that can be differentiated based on ecological pressures and/or specific conditions of morbidity, such as the elderly, bedridden patients and patients with chronic diseases, particularly diabetes mellitus. Studies suggest that the rate of nasal colonization with S. aureus and MRSA may be higher in diabetic patients than in non-diabetic subjects.35,39,40 Within this context, an influence of glycemic control on the S. aureus carriage rate has been demonstrated, considering that hyperglycemia reduces the activation of macrophages.39,40 These patients are also more susceptible to persistent infections, particularly skin infections such as those caused by S. aureus, and are more likely to develop severe MRSA pneumonia due to increased blood glucose levels and suppression of the immune response.41,42 A prospective cohort study showed that patients with both type 1 and type 2 diabetes mellitus had an increased risk of bacterial skin infections; a higher risk of recurrence is observed in patients with type 1 diabetes, which can be explained by the higher frequency of nasal carriage observed among insulin-dependent diabetic patients.43 A recent study conducted by our group evaluated nasal and oropharyngeal MRSA carriage in patients with insulin-dependent diabetes from Botucatu and found a 30.4% prevalence of colonization with S. aureus and a 4.8% prevalence of colonization with MRSA.44 The S. aureus colonization rate was very similar to the overall prevalence found in the population of the same city in a previous study.34 However, MRSA isolates were more frequent among diabetic patients (4.8% vs 0.9%).
The emergence of CA-MRSA also poses a special risk to known vulnerable populations such as the elderly, especially residents of long-term care facilities. Within this context, the prevalence of colonization with S. aureus and MRSA among elderly residents of nursing homes and bedridden patients from Botucatu and Bauru (interior of São Paulo) was investigated by two different studies. In Bauru, the prevalence of S. aureus and MRSA nasal carriage was 17.7% and 3.7%, respectively.45 On the other hand, a higher colonization rate was observed in Botucatu, which reached 33.6% for S. aureus and 8% for MRSA, probably because the analysis included extra-nasal sites (oropharynx and anal).46
Long-term care facilities are intermediate spaces between the community and the hospital and may represent a possible reservoir of multidrug-resistant microorganisms.47 Furthermore, the social and interactive characteristics of nursing homes, such as shared meals, recreation and therapeutic facilities, certainly facilitate the spread of communicable diseases.48 However, little is known about the prevalence of MRSA among older adults, with rates ranging from 0.7 to 2.0% in studies conducted around the world that are in contrast to the above-mentioned findings.49 In particular, a recent study investigating nasopharyngeal colonization in 776 older outpatients attending the largest geriatric clinic in the city of São Paulo found a prevalence of S. aureus and MRSA carriage of 15.9% and 2.3%, respectively.49 Here it is important to note that nursing homes in Brazil can be public, private or philanthropic, and the quality of care varies widely, a fact that influences the spread of pathogens.45
As reported for diabetic patients in the aforementioned study,44 SCCmec characterization of MRSA isolated from older adults living in the same region showed a predominance of SCCmec types I, II and IV,45,46 with SCCmec I and II occasionally being risk factors for HA-MRSA. Similar SCCmec typing results were obtained for MRSA isolated from patients of a psychiatric hospital and from outpatients with skin infections treated at a dermatology clinic in the same city.50,51
Another interesting study drew attention to the circulation of strains that were resistant to different classes of antimicrobial agents in the community, demonstrating a high prevalence of S. aureus (51.5%) and MRSA (8.7%) isolated from patients with skin wounds treated at primary healthcare centers in Botucatu, with no reports of risk factors for HA-MRSA. The strains carried SCCmec II and IV and a positive association was found between nasal carriage of S. aureus and MRSA and their respective presence in the wound.52 However, the gene encoding PVL was not detected, a finding that is consistent with the low prevalence of this gene in MRSA-IV reported in Brazil.52
Clonal diversity of CA-MRSA and epidemiological changesStudies investigating the clonal profile of CA-MRSA isolates throughout Brazil identified strains predominantly characterized as SCCmec IV, including those that cause staphylococcal infections; special attention must be paid to clone ST5-IV whose frequent identification suggests its spread, particularly in Botucatu and region. These and other studies on the clonal characterization of MRSA isolated from the community in Brazil are described in Table 2, which provides an overview of CA- MRSA clones in Brazil reported in the literature.
Community-associated MRSA clones in Brazil reported in the literature.
| Clonal complex | Sequence type | SCCmec type | Study location (city) | References |
|---|---|---|---|---|
| CC5 | ST5 | IV | Botucatu, Avaré, Bofete, São Paulo, Vitória da Conquista, Pardinho | 27,34,44,53,57,66,76,77 |
| I | Botucatu | 44 | ||
| II | Botucatu | 57 | ||
| ST2594 | IV | Botucatu | 34 | |
| ST1176 | IV | Botucatu, Fartura | 34,57,66 | |
| ST6 | IV | Botucatu | 66 | |
| CC8 | ST8 | IV | Botucatu, Porto Alegre, Paranapanema | 44,66,57,14,25,66 |
| ST239 | III | Goiânia | 55 | |
| CC45 | ST1120 | V | Goiânia | 55 |
| ST45 | IV | Porto Alegre, Vitória da Conquista | 25,53 | |
| ST2228 | IV | Vitória da Conquista | 53 | |
| CC1 | ST1 | IV | Rio de Janeiro, Porto Alegre | 14,20 |
| CC30 | ST30 | IV | Porto Alegre, Rio de Janeiro, Goiânia, Botucatu | 13,14,79,55,80,25,66 |
| CC121 | ST121 | IV | Goiânia | 55 |
| Singleton | ST12 | IIIa | Goiânia | 55 |
CC: Clonal complex; ST: Sequence type.
Reports of isolates carrying SCCmec type V are less common in Brazil, and SCCmec III isolates are usually multidrug-resistant and associated with HAIs.53,54 However, both types were found colonizing the nares of healthy children attending day care centers.53,55 In fact, strains carrying SCCmec III seem to be adapting to the community and are no longer restricted to the hospital.55 SCCmec III MRSA isolates have also been described colonizing health professionals working in primary healthcare centers in the inland of the country, including community health agents who perform tasks that require a greater level of contact with the population.56 Like SCCmec III, SCCmec types I and II are commonly found circulating in health services, including primary healthcare settings, as reported by Pereira-Franchi et al.52,57 and Goes et al.56; a history of hospitalization is frequently associated with this finding.
Interesting findings from a study conducted in a hospital in the interior of São Paulo indicate polyclonal endemicity, with hegemony of SCCmec III but unrelated to the Brazilian endemic clone, and suggest wide dissemination of MRSA in Brazilian hospitals, in which SCCmec IV isolates are acquired in the healthcare settings.58 In fact, several epidemiological studies have reported blurring of boundaries between CA-MRSA and HA-MRSA, indicating a significant overlap between the two groups.59 Nevertheless, SCCmec typing continues to be an important tool to elucidate the evolution of MRSA, as well as to understand and monitor constant epidemiological changes and to guide therapeutic decisions. The concept of SCCmec elements should be revised considering their notorious ability to carry other genes that are essential for the increased survival of staphylococci in different environments.11,59
A study conducted in Bahia identified a high prevalence of nasal colonization with CA-MRSA (47.3%) in healthy children attending public day care centers, with the description of lineages belonging to CC5 (ST5), CC45 (ST45 and ST2228) and CC398 (ST389) that carry SCCmec IV and V.53 Interestingly, the ST398 clone, which is traditionally associated with livestock, has emerged among human patients, including in Brazil, and will be discussed below. Complexes CC5 and CC45 belong to clonal groups involved in a global pandemic caused by MRSA; however, CC5 lineages are notably the most frequently detected in Brazil, especially ST5. Other clones included in CC5 have also been reported in Brazil, such as ST2594, ST1176 and ST6. The ST1176 clone was described for the first time in S. aureus SCCmec IV of patients admitted to a hospital in the city of São Paulo and was identified in Botucatu (140 miles away from São Paulo) by two community studies; one of these studies also identified a new clone, ST2594, circulating in the city.34,57,60
Reports of CA-MRSA are less common in European countries where infections with MRSA are typically healthcare associated.61 Characteristically, there is an extensive clonal diversity of CA-MRSA in Europe, with a high degree of geographic segregation of clones, associated with a considerably low but rising prevalence in some countries.62 In general, the European clone ST80-MRSA-IV (lukSF-PV positive) predominates, although the USA300 clone (ST8-MRSA-IV) has been reported throughout the United Kingdom and Europe.62 Greece in particular is the country with the highest incidence of CA-MRSA in Europe.63 On the other hand, CA-MRSA infections have emerged in Nordic countries and in The Netherlands, where rates of HA-MRSA are very low.61 An important epidemiological aspect in The Netherlands is the emergence of clone ST398-V, which is responsible for about 20% of cases of MRSA infection in the country.61,62
Clone ST398Described for the first time in France, the CC398-ST398 lineage originated in animals and is one of the most important genotypes of livestock-associated S. aureus (LA-MRSA). First documented in countries with intensive animal farming (cattle, horses, pigs), these microorganisms are increasingly spreading among humans not exposed to these animals.8 Human infections with the ST398 clone have sporadically been reported in various geographic regions, including Europe, the Americas and Asia, with a higher prevalence in Europe and China.64
Cases of human colonization and infection with methicillin-susceptible S. aureus (MSSA)-CC398 appear to be more common, whereas MRSA-ST398 strains are mainly detected in animals due to clonal adaptation.65 These strains are generally not associated with the production of PVL or enterotoxins, but exhibit a remarkable diversity of resistance genes, including resistance to trimethoprim, tetracycline, macrolides, lincosamides, gentamicin, ciprofloxacin and trimethoprim-sulfamethoxazole, as well as to antibiotics used in livestock farming.7,8 Other important features of this emerging clone include its ability to acquire virulence genes, low host specificity, and skilled mobility and spread, characteristics that render this lineage a real threat.7
In Brazil, asymptomatic colonization with the ST398 clone was identified in MSSA and MRSA isolated from diabetic patients (MSSA/MRSA-ST398-IV)44 and people living with HIV/AIDS (MSSA-ST398)66 in Botucatu/SP, healthy children in Vitória da Conquista/BA (MRSA-ST398-V),53 and children attending day care centers and public hospitals in Niterói/RJ (MRSA-ST398-IV/V). The genes encoding PVL were detected in these isolates.67 These findings indicate that the ST398 clone might be emerging in the community in different parts of the country and in different population groups, with this occurrence being underreported, as suggested by Neto et al.67
The ST398 clone has also occasionally been reported to cause severe infections in Brazil. A prospective study conducted at the Hospital of the Botucatu Medical School revealed a considerable number of MSSA isolates that belonged to the ST398 clone and that were associated with cases of pneumonia and a higher mortality rate.68 For example, Gales et al.69 reported a case of fatal pneumonia caused by MSSA-ST398 in a cancer patient from São Paulo.69
MRSA and COVID-19The current circumstances of the COVID-19 pandemic, as well as of previous pandemics, raise important issues that need to be investigated. Studies show that about half of the patients hospitalized with COVID-19 who die are co-infected with fungi and bacteria.70,71 The widespread use of antibiotics in the treatment of these bacterial co-infections highlights the importance of considering possible effects on the global prevalence of antibiotic-resistant bacteria.72
During an outbreak of the so-called severe acute respiratory syndrome (SARS-CoV-1) in 2003, an increased frequency of MRSA transmission and infection was reported among intensive care unit patients, which was strongly linked to ventilator-associated pneumonia.73 Furthermore, bacterial pathogens, including S. aureus, have been recognized to be involved in infections secondary to influenza and are a common cause of post-influenza bacterial pneumonia. Within this context, the emergence of CA-MRSA strains in recent decades has changed the clinical scenario of these infections.74
Certainly, the current pandemic scenario will lead to significant changes in the pattern of endemic pathogens such as S. aureus. Studies highlight the complex and multifaceted interrelationships and interdependence of antimicrobial resistance determinants in past, current and future pandemics. Potential interventions to support the reduction of antimicrobial prescriptions during the COVID-19 pandemic require urgent consideration.75
ConclusionWe understand that a broad and simultaneous approach to the special populations described above, combining epidemiological strategies and the genetic characterization of staphylococci, will provide valuable information. We also believe that the evaluation of different groups will provide insights into the pathogenesis and spread of MRSA strains.
Cases of CA-MRSA colonization have been described in all regions of Brazil; however, the use of molecular typing techniques that provide more refined data on the epidemiology of these pathogens in Brazil is still limited. In addition, available data on CA-MRSA infections in Brazil are mainly derived from case reports. It is nevertheless possible to catch a glimpse of aspects of the epidemiology of these pathogens in the country, recognizing possible reservoirs in the community, especially children attending day care centers, institutionalized older adults, and diabetic patients.
On the other hand, some studies have reported the lack of an epidemiological link between the type of SCCmec and a history of hospital admission, corroborating the current global scenario of the widespread transmission of MRSA isolates between the community and hospitals. All of these points, together with the lack of epidemiological surveillance of S. aureus and MRSA in Brazil, highlight the importance of the adoption of assertive strategies by all government sectors and by society, with a reassessment of current public policies in light of the new perspectives arising from epidemiological findings.
We thank the National Council for Technological and Scientific Development (CNPq; Grant 303603/2020-8) and PROPG Edital 19/2021.