Nontuberculous mycobacteria (NTM) species are increasingly being isolated and have become a key factor affecting public health by causing pulmonary diseases. Most NTM species do not respond to conventional tuberculosis (TB) drugs. This study aimed to identify NTM isolated from suspected pulmonary TB patients from the Zhejiang province and analyze their distribution in the region.
MethodsA total of 1,113 NTM isolates from patients suspected to be suffering from acid-fast bacilli-positive tuberculosis were identified at the species level, using the CapitalBio Mycobacterium identification array and polymerase chain reaction amplification and sequencing of 16S-23S gene internal transcribed spacer (ITS), 16S rRNA, and hsp65.
ResultsOf the 23,138 isolates, we identified 1,102 NTM (4.8%), mainly including Mycobacterium intracellulare (54.81%, 604/1,102), M. chelonae-M. abscessus (16.52%, 182/1,102), M. avium (13.16%, 145/1,102), M. kansasii (8.17%, 90/1,102), and M. gordonae (3.27%, 36/1,102).
ConclusionThe distribution of NTM species observed in patients with suspected pulmonary tuberculosis provides guidance for the diagnosis and treatment of NTM pulmonary diseases.
Nontuberculous mycobacteria (NTM) refer to Mycobacterium species other than M. tuberculosis complex (MTBC) and M. leprae.1 NTM are opportunistic pathogens of animals and humans that exist in the environment.2 Most NTM cause pulmonary diseases, which usually occur in underlying structural airway diseases such as bronchiectasis or chronic pulmonary disease.3 In recent years, in many countries, the rate of NTM isolation from respiratory specimens and the number of patients infected with NTM have been increasing rapidly.4-6 Importantly, most NTM (except M. kansasii) are resistant to all commonly used anti-tuberculous drugs or susceptible to only partially standard anti-tuberculous drugs.7 More and more people realize the importance of NTM diagnosis in the treatment of lung diseases.8 Thus, the identification of NTM species in respiratory specimens is critical for choosing effective regimen.
Among many known NTM species, only a small number cause pulmonary disease in humans. The most common slowly growing mycobacteria (SGM) are members of M. avium complex (MAC),9 including M. avium and M. intracellulare. Among rapidly growing mycobacteria (RGM), M. abscessus complex (MABC), M. chelonae and M. fortuitum are by far the most common pathogens of pulmonary disease. The pathogenicity of NTM varies greatly among different species. For example, M. gordonae is considered to rarely cause disease in humans, while M. kansasii is pathogenic.10 For low pathogenic species, such as M. gordonae, several repeated tests for the presence of positive cultures within a few months, as well as strong clinical and radiological evidence of the disease, would be necessary to determine whether M. gordonae caused the disease. On the other hand, for M. kansasii isolated, a positive culture may be sufficient to support the initiation of antimicrobial treatment.11
In China, tuberculosis (TB) is still a serious public health problem. Therefore, some patients with the sputum specimens tested positive for acid-fast bacilli (AFB) would be misdiagnosed as pulmonary TB and treated with anti-TB drugs. In China, people lack comprehensive understanding of the degree of misdiagnosis and inappropriate treatment of disease due to NTM infection. However, NTM isolation does not necessarily mean disease diagnosis, once NTM may be contamination or colonization. Thus, when facing the isolation of NTM species, clinicians have difficulty in diagnosing NTM infection.12 Therefore, the correct identification of NTM must be performed to provide an appropriate treatment regimen.13
In the present study, we collected sputum specimens from patients with suspected pulmonary TB in a longitudinal study (2009–2019) from 49 hospitals or TB surveillance stations in Zhejiang province. Furthermore, we analyzed the distribution of the isolated NTM species in the province.
Material and methodsStrainsA total of 23,138 isolates from patients with suspected AFB-positive tuberculosis were collected from 49 hospitals or TB surveillance stations in 2009 to 2019 and transported to the Center for Disease Control and Prevention in Zhejiang province, China. All isolates were cultivated on Löwenstein-Jensen medium. Ethical approval was granted by the Center for Disease Control and Prevention in Zhejiang province Research Ethics Committee (Ethics number: 2017-005). Data collection was conducted under regulations covering information collection.
Immunochromatographic identification using MPB64-ICAThe MPB64-ICA (Hangzhou Genesis Biodetection & Biocontrol Co., Ltd., Hangzhou, China) is a rapid immunochromatographic identification test for the M. tuberculosis complex that uses anti-MPB64 monoclonal antibodies. The test was performed according to the manufacturer's instructions strictly.14 In brief, 100 μL of broth cultures were inoculated to the 96-well plates and then allowed to stand at 25°C for 15 min. A red band in the test zone is produced if MPB64 is present in the sample (Fig. 1).
DNA extractionNTM colonies were recovered from sputa of patients with suspected pulmonary TB and scraped onto 7H10 medium (Becton, Dickinson and Company, USA). Genomic DNA was extracted using the DNeasy Blood & Tissue Kit (QIAGEN, Valencia, CA, USA), following the manufacturer's protocol as previously reported.15
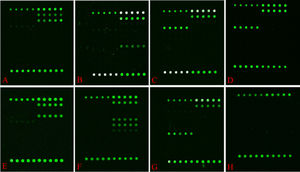
Detection using CapitalBio Mycobacterium identification array systemDNA microarray chip from Mycobacterial Species Identification Array Kit (CapitalBio Technology Inc., Beijing, China) was performed for NTM species identification as previously reported.16-18 It can identify as many as 17 Mycobacterium species: M. tuberculosis complex, M. chelonae-M. abscessus, M. fortuitum, M. intracellulare, M. avium, M. kansasii, M. gordonae, M. terrae, M. smegmatis, M. szulgai/M. malmoense, M. nonchromogenicum, M. scrofulaceum, M. xenopi, M. aurum, M. marinum/M. ulcerans, M. gilvum, and M. phlei (Fig. 2).
Hybridization patterns of representative strains on the probe array (Mycobacterial Species Identification Array Kit (CapitalBio Technology Inc., Beijing, China)). Green fluorescence indicates positive hybridization to the probe. M. intracellulare (A), M. chelonae and M. abscessus (B), M. avium (C), M. kansasii (D), M. gordonae (E), M. fortuitum (F), M. terrae (G), Negative control (H).
The PCR amplification of 16S-23S rRNA gene internal transcribed spacer (ITS), 16S rRNA, and hsp65 were performed. Primers and the PCR cycling conditions are listed in Table 1. PCR products were purified and sequenced on ABI3730XL (Shanghai Shenggong Biological Engineering Technology Co., Ltd. Shanghai, China). The consensus 16S-23S ITS, 16S rRNA, and hsp65 sequences of each isolate were analyzed using BLAST.19 Sequence similarity alignment referred to the microbial database of representative genome, and only highly similar sequences were selected, while other settings were set by default. The identification criteria for sequence similarity at the species level using the database is ≥ 97%.
Primers used in this study.
| Gene | Direction* | Sequence (5′3′) | Conditions |
|---|---|---|---|
| 16S-23S ITS | F | GGG TAC TGA GAT GTT TCA CTT C | 95°C for 5 min |
| R | TCA CCT CCT TTC TAA GGA GCA CC | 35 cycles (94°C for | |
| 16s rRNA | F | AGT TTG ATC CTG GCT CAG | 35 s; 64/56/60°C for 30 s; 72°C for 50 s) |
| R | GGT TAC CTT GTT ACG ACT T | ||
| hsp65 | F | ACC AAC GAT GGT GTG TCC AT | |
| R | CTT GTC GAA CCG CAT ACC CT | 72°C for 10 min |
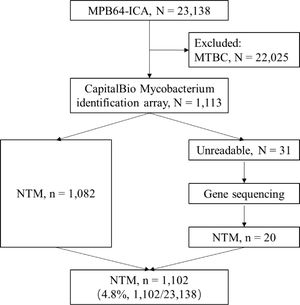
Of the 23,138 isolates different patients with pulmonary diseases, 22,025 were identified as MTBC by MPB64-ICA positive, while the remaining 1,113 were identified as non-MTBC isolates. The non-MTBC isolates were then subjected to testing using the CapitalBio Mycobacterium identification array. Among the isolates, 97.21% (1,082/1,113) were identified at species level using positive hybridization with the NTM-specific probes, whereas 2.79% (31/1,113) isolates were identified as unreadable results using the CapitalBio array assay (Fig. 3). Then, all 31 unidentifiable mycobacterium isolates were sequenced and identified at species level using BLAST. As a result, 20 NTM and 11 non-mycobacteria isolates were identified. In total, we obtained 4.8% (1,102/23,138) NTM isolates from 23,138 patients with suspected acid-fast bacilli-positive tuberculous (Fig. 3).
The spectrum of NTM speciesAs presented in Table 2, 23 NTM species were detected. Among them, 13 were detected using the CapitalBio assay and 10 species were detected through gene sequencing. The five most dominant species out of 1,102 NTM isolates accounted for 95.93%, which were M. intracellulare (54.81%, 604/1,102), M. chelonae-M. abscessus (16.52%, 182/1,102), M. avium (13.16%, 145/1,102), M. kansasii (8.17%, 90/1,102), and M. gordonae (3.27%, 36/1,102). Other NTM isolates accounted for 4.07% in total, mainly including M. fortuitum (0.82%, 9/1,102), M. scrofulaceum (0.45%, 5/1,102), M. peregrinum (0.45%, 5/1,102), M. intermedium (0.36%, 4/1,102), M. terrae (0.27%, 3/1,102) (Table 2). Particularly, one M. tuberculosis and M. intracellulare mixed infection was detected using the array assay.
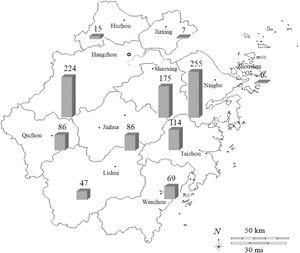
NTM distribution among pulmonary NTM patients, Zhejiang province, 2009-2019.
As presented in Fig. 4, 1084 NTM isolates have been detected in 11 major cities in the Zhejiang Province. Unfortunately, we could not determine the origin of 18 NTM strains out of 1,102 isolates. The six most prevalent cities were Ningbo (23.52%, 255/1,084), Hangzhou (20.66%, 224/1,084), Shaoxing (16.14%, 175/1,084), Taizhou (10.52%, 114/1,084), Quzhou (7.93%, 86/1,084), and Jinhua (7.93%, 86/1,084), which accounted for 86.72% of all NTM identified. Other cities, including Wenzhou (6.37%, 69/1,084), Lishui (4.34%, 47/1,084), Huzhou (1.38%, 15/1,084), Jiaxing (0.65%, 7/1,084), and Zhoushan (0.55%, 6/1,084), accounted for only 13.28% of the isolates.
DiscussionNTM can lead to symptoms that are difficult to distinguish from those of TB, and pose challenges to diagnosis and treatment, especially in low-income and middle-income countries or regions.20 NTM accounts for the majority of the suspected pulmonary TB cases in the world. The proportion of NTM in suspected pulmonary TB was approximately from 4.2% to 6.4% in mainland China. In our study, there were 4.8% (1,102/23,138) NTM isolates among suspected pulmonary TB cases from 49 hospitals or TB surveillance stations from 2009 to 2019 in Zhejiang province. In a national study, there were 6.4% (315/4,917) NTM isolates among mycobacterial isolates from 72 nationwide TB surveillance sites in 31 provinces of mainland China in 2013.21 In a study in Fujian province, China, 1,425 isolates were included, of which 60 (4.2%) were identified as NTM species from July 1, 2010 to June 30, 2011.22 However, the proportion of NTM in suspected pulmonary TB varies greatly between different countries or regions: 0.6% (60/10,466), 2.5% (43/2,036), 7.1% (68/958) and 15.1% (62/410) in Northern Tunisia, Ghana, Korea and Iran, respectively.13,20,23,24 Thus, the species distribution of NTM isolates and the types of diseases caused by NTM species vary per region.25,26
MAC is the most frequently isolated NTM species from infected pulmonary samples in southeast China,22,27 Japan,28 the United States of America,29 and Ghana.13 Isolation frequency between MAC species is observed to be different per continent.25 Especially, M. intracellulare is the most frequently isolated species in some countries (Table 3). Majority of MAC isolates are causative agents of clinically relevant diseases, that is, the patient ultimately meets the American Thoracic Society/Infectious Disease Society of America diagnostic criteria for NTM pulmonary disease. MAC pulmonary disease typically has two major clinical types: the nodular bronchiectatic form and fibrocavitary form.30
The comparison of NTM distribution of pulmonary cases from various studies.
| District/nation | year | NTM proportion | Species rank | methods | source |
|---|---|---|---|---|---|
| China, Zhejiang province | 2009-2019 | 4.8% (1,112/23,138) | M. intracellulare (54.81%); M. chelonae and M. abscessus (16.52%); M. avium (13.16%); M. kansasii (8.17%) | CapitalBio Mycobacterium identification assay | This study |
| China | 2013 | 6.4% (317/4,917) | MABC (36.0%); MAC (34.1%); M. kansaii (9.8%); M.paragordonae (5.4%) | MALDI-TOF MS | 21 |
| China, Fujian province | 2011 | 4.2% (60/1,425) | M. intracellulare (68.3%); M. abscessus (10.0%); M. avium (6.7%); M. kansasii (6.7%); | 16S rRNA, hsp65, rpoB, 16S-23S rRNA ITS | 22 |
| China, Guangdong province | 2013-2016 | - | MAC (44.5%); MABC (40.5%); M. kansasii (10.0%); M. fortuitum (2.8%) | - | 27 |
| Japan | 2006-2016 | - | MAC (87.3%); MABC (5.5%); M. kansasii (3.9%) | - | 28 |
| America | 2005-2015 | - | MAC (83.2%); M. kansasii (7.7%); M. abscessus (3.4%) | - | 29 |
| Northern Tunisia | 2019 | 0.6% (60/10,466) | M. kansasii subtype 1(23.3%); M. fortuitum (16.6%); M. novocastrense (16.6%); M. chelonae(10.0%); | rpoB, 16S rRNA, hsp65, sodA | 23 |
| Ghana | 2017 | 2.5% (43/1,755) | M. intracellulare(41.9%); M. avium subsp paratuberculosis (30.2%); M. abscessus (11.3%); M. mucogenicum (7.0%) | IS6110, hsp65 | 13 |
| Korea | 2009 | 7.1% (68/958) | M. abscessus (31%); M. fortuitum (15%); M. avium complex (8%); M. gordonae (8%); | rpoB | 24 |
| Iran | 2018 | 15.1% (62/410) | M. simiae (38.7%); M. fortuitum (19.3%); M. kansasii (17.7%); M. avium complex (8.0%); | 16S rRNA, rpoB, hsp65, ITS | 20 |
Another NTM species that shows a remarkable difference in isolation frequency is MABC, which is common in Korea,24 China,21,22,27 Japan,28 USA,29 and Ghana13 but less frequent Northern Tunisia23 and Iran.20 MABC is a RGM that is responsible for opportunistic pulmonary infections in patients with structural lung disorders, such as cystic fibrosis and bronchiectasis, as well as a wide range of skin and soft tissue infections in humans.31 In addition, MABC is one of the most severe drug resistant bacteria among the RGM; therefore, it is very difficult to treat.32
It is worth noting that M. kansasii is also commonly isolated in many countries or regions. Remarkably, the isolation rate of this pathogen from NTM pulmonary diseases in Norther Tunisia was particularly high, reaching 23.3% of total NTM isolations,23 as compared with the observed rates in Japan (3.9%)28 and USA (7.7%).29 Pulmonary disease caused by M. kansasii tends to cluster in specific geographical locations, with Central Europe being one of the hotspots.33 However, since M. kansasii has been specially recovered from municipal tap water,34 its distribution in different areas may be attributed to the municipal water systems rather than specific ecological environment.35 Typically, disease caused by M. kansasii is characterized by apical fibro-cavitary lung disease, which is very similar to pulmonary TB clinically. Another uncommon pulmonary manifestation is nodular bronchiectasis. Extrapulmonary manifestations are even rarer, including lymphadenitis, skin and soft-tissue infections, and disseminated disease.35
Correct identification of NTM is important, as it can predict the clinical relevance of an isolate as well as aid in the selection of a treatment regimen. Both molecular and mass spectrometry-based methods can be applied. Molecular identification is the preferred method, and it can be achieved by using probes or through gene sequencing. Probe-based assays are easier to perform and implement. Meanwhile, gene sequencing allows a higher level of discrimination, often up to subspecies level, but it is only feasible for laboratories with access to sequencing facilities. Thus, the probe-based assays are easier to popularize.
Microarrays are a valuable tool for the specific detection of microorganisms directly from clinical samples. Compared with other assays, the CapitalBio Mycobacterium identification array system shows higher sensitivity and specificity.36 It has been reported that the sensitivity and specificity of the CapitalBio Mycobacterium identification microarray system were 99.6% and 100%, respectively, compared with the results of 16S rRNA gene sequencing when used with clinical isolates from respiratory specimens.16 Interestingly, it can identify up to 17 Mycobacterium species. The microarray method can detect more than 98% of the pathogens of pulmonary diseases. Thus, only a few mycobacterium pathogens cannot be identified. Therefore, the identification method we have been using for 10 years is reliable and is significant in rapid NTM classification, understanding of its prevalence, and clinical diagnosis and treatment.
This study has several limitations. First, the array method cannot identify M. chelonae-M. abscessus, M. marinum/M. ulcerans, and M. szulgai/M. malmoense at species level. Second, our study is a descriptive study; we did not perform gene sequencing at the same time to verify the accuracy of the microchip method. Third, the sociodemographic, clinical and radiological data are not detailed available, as the NTM isolates were collected from many hospitals in the province and transported to the Center for Disease Control and Prevention in Zhejiang province. The pathogens of NTM pulmonary diseases were collected and identified to species level to provide guidance for clinical treatment. Forth, in this study, although at least two sputum specimens for positive culture were collected for all patients, only one sample was sent to us for identification. Therefore, most patients did not have two consistent identifications for NTM, so in the strict sense, not all patients were identified as NTM pulmonary disease, since the guide on the diagnosis and treatment of NTM disease in China requires two sputum specimens collected separately were positive for NTM culture and identified as the same pathogen.37,38
In conclusion, the misdiagnosis of NTM among suspected pulmonary TB patients has highlighted the urgent need for sensitive, reliable, and fast methods for the laboratory detection of mycobacteria. This study demonstrates that the CapitalBio Mycobacterium identification array system is a reliable and rapid approach for the identification of the most frequently isolated mycobacteria. The distribution of NTM can provide a reference for the diagnosis and treatment of NTM pulmonary diseases.
FundingThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
None.