Listeria monocytogenes is responsible for causing listeriosis, a type of food poisoning with high mortality. This bacterium is mainly transmitted to humans through the consumption of contaminated foods. Detection of L. monocytogenes through molecular methods is crucial for food safety and clinical diagnosis. Present techniques are characterized by low discrimination power and high cost, as well as being time-consuming and taking several days to give the final result. In our study, MLVA-HRM (Multiple-Locus Variable-number tandem repeats Analysis ‒ High-Resolution Melting) was investigated as an alternative method for a fast and precise method for the genotyping of L. monocytogenes isolates. Forty-eight isolates of L. monocytogenes obtained from the microbial bank of Department of Microbiology, Iran University of Medical Sciences, were typed by MLVA-HRM analysis using five Variable Numbers of Tandem Repeat (VNTR) loci. A total of 43 different types were obtained. This research demonstrated the usefulness of the MLVA-HRMA method and its ability to discriminate L. monocytogenes isolates. Since this method is easier and more efficient than existing methods, it can be widely used in food processing plants and diagnostic laboratories as a fast and accurate method.
Listeria monocytogenes is a Gram-positive, motile, non-spore forming, and food-borne pathogeindustry, an cause listeriosis in high-risk individuals, including infants, the elderly, and immunocompromised patients.1,2L. monocytogenes is widely distributed in the environment and has the ability to survive and grow in harsh conditions, such as low temperatures and high saline concentrations.3L. monocytogenes is an environmental organism that typically infects food in the processing industry and it is estimated that 99% of human listeriosis infections are caused by the consumption of contaminated foods.4 Listeriosis is linked with a high rate of hospitalization (85%–90%) and the mortality rate for this disorder is 20%‒30%, which is higher than infections caused by other food poisoning pathogens.5,6
Thus, in recent decades, L. monocytogenes has become a major concern in the food industry and public health, and the identification of L. monocytogenes species for food safety, epidemiological studies, and clinical diagnosis became very critical.7
There are several methods for the classification of bacterial isolates, such as Multilocus Sequence Typing (MLST), Pulsed-Field Gel Electrophoresis (PFGE), and restriction enzyme analysis.8,9
Although each of these methods has its advantage in isolating different bacterial species, the typing technique should be simple and easy. PFGE is the current gold standard for typing of L. monocytogenes isolates, however, it is time-consuming and hard to standardize, which prevents the exchange of typing results between laboratories.10 The Multiple-Locus Variable-number tandem repeats Analysis (MLVA) method requires electrophoresis of PCR products and the MLST method requires a large sequence analysis.11,12 Therefore, considering the importance of typing food-related bacteria, it is very crucial to use a fast, cheap, and easy method to differentiate the strains.13
High-Resolution Melting (HRM) is a Quantitative PCR (QPCR)-based method developed to detect changes in nucleic acid sequences. This method monitors changes in DNA sequences according to the changes of melting temperatures of real-time PCR products.14 Through our research, five Variable Numbers of Tandem Repeat (VNTR) loci were selected for the genotyping of L. monocytogenes isolates. VNTR is a region in DNA where a short nucleotide sequence called Tandem Repeats (TRs) with various numbers are located in different strains of bacteria.15,16 Today, this difference in the number of VNTR is used as a suitable target for assessing bacterial genotyping.17 In this study, we used the HRM method, which is a simple and fast method for the analysis of VNTR and genotyping of L. monocytogenes strains.
Materials and methodsBacterial strains and growth conditionsIn this study, L. monocytogenes ATCC 19115 and 47 clinical L. monocytogenes strains from the Microbial bank of Department of Microbiology affiliated to Iran University of Medical Sciences, Tehran, Iran, were used (Table 1).18 Isolates were cultured on Brain Heart Infusion (BHI) agar (Merck, Darmstadt, Germany) at 37°C for 48h. DNA of the samples was extracted with a DNA Extraction kit (Roche, Germany) according to the manufacturer's protocol. DNA purity and concentration were assessed using a NanoDrop Spectrophotometer (Thermo Scientific, USA).
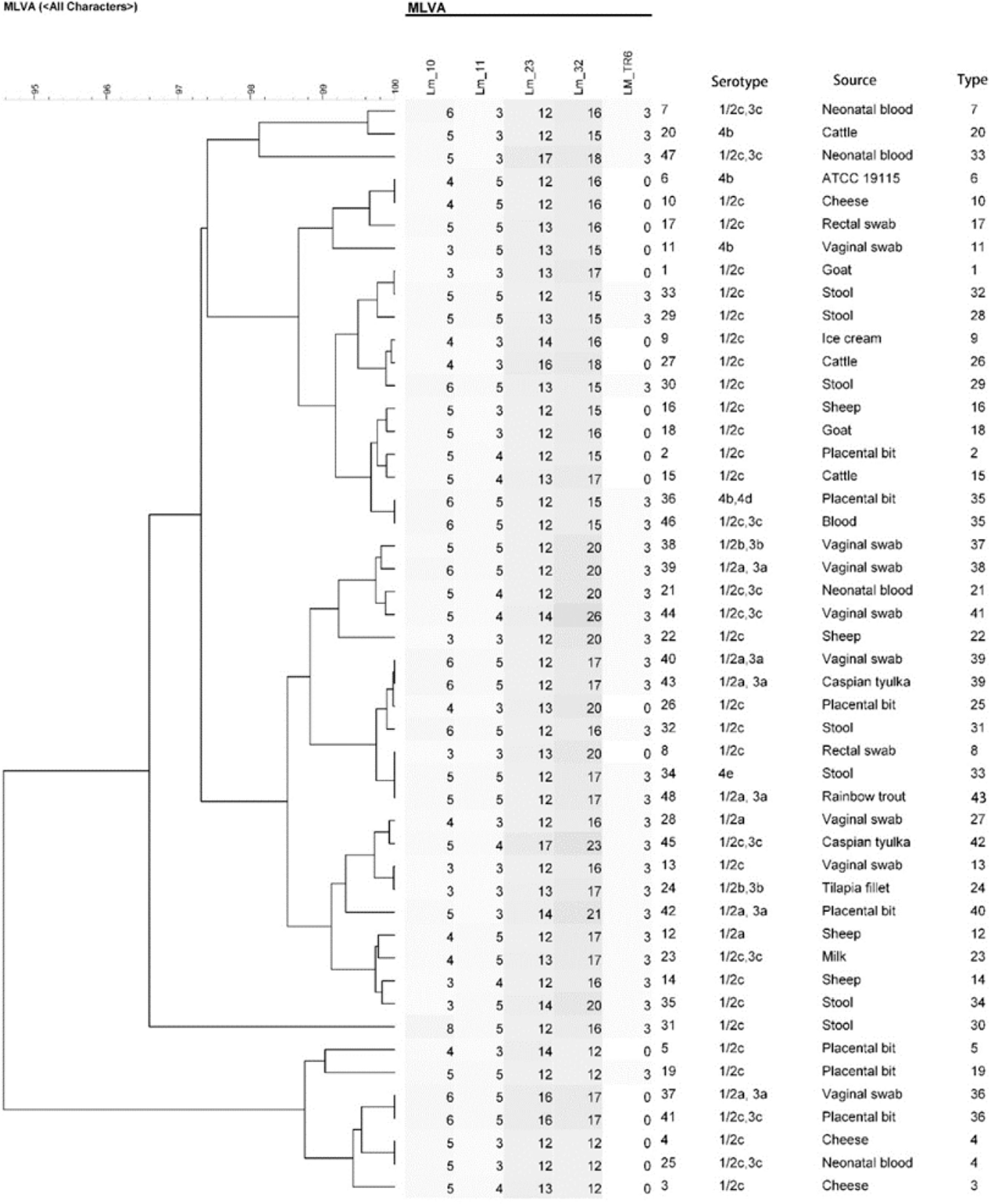
Serotypes and sources of Listeria monocytogenes strains.
In this study, we used 5 VNTR loci for MLVA-HRMA based on the previously designed primers.19 The PCR amplification and HRMA were performed using a Rotor-Gene thermal cycler (Corbett Life Sciences, Sydney, Australia). The PCR reaction solution was performed with a total of 20 µL, containing 1 µL of each forward and reverse primers, 1 μL of template DNA (0.5 μg), 4 μL of 5× Hot Firepol Eva Green HRM Mix (Solis BioDyne), and 13 μL of sterile distilled water.
The thermal program was performed by an initial denaturation at 95°C for 5 min followed by 40 cycles of 95°C for 30s, 54°C for 22s, and 72°C for 40s for VNTR locus 1 (Lm_10), 95°C for 30s, 52°C for 22s, and 72°C for 40s for VNTR locus 2 (Lm_11), 95°C for 30s, 55.5°C for 30s, and 72°C for 30s for VNTR locus 3 (Lm_23), 95°C for 30s, 54°C for 30s, 72°C for 45s for VNTR locus 4 (Lm_32) and 95°C for 30s, 57°C for 22s, and 72°C for 40s for VNTR locus 5 (LM_TR6).
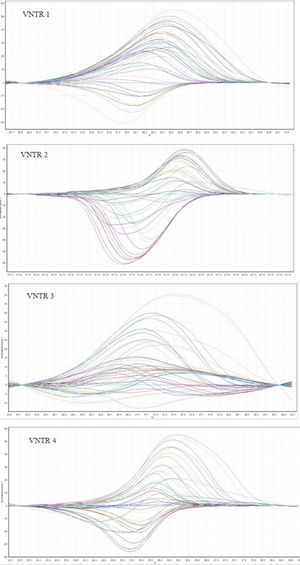
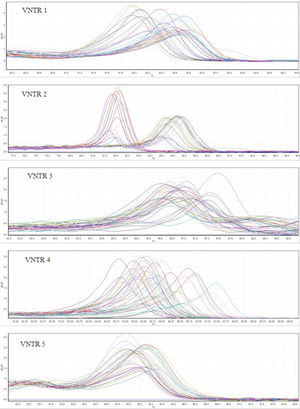
After PCR amplification, the HRM step was performed. For HRM step the fluorescence was measured by increasing the temperature from 75° to 95°C with a rate of 0.1°C/s. In case of nucleotide changes in different strains, various curves were produced.
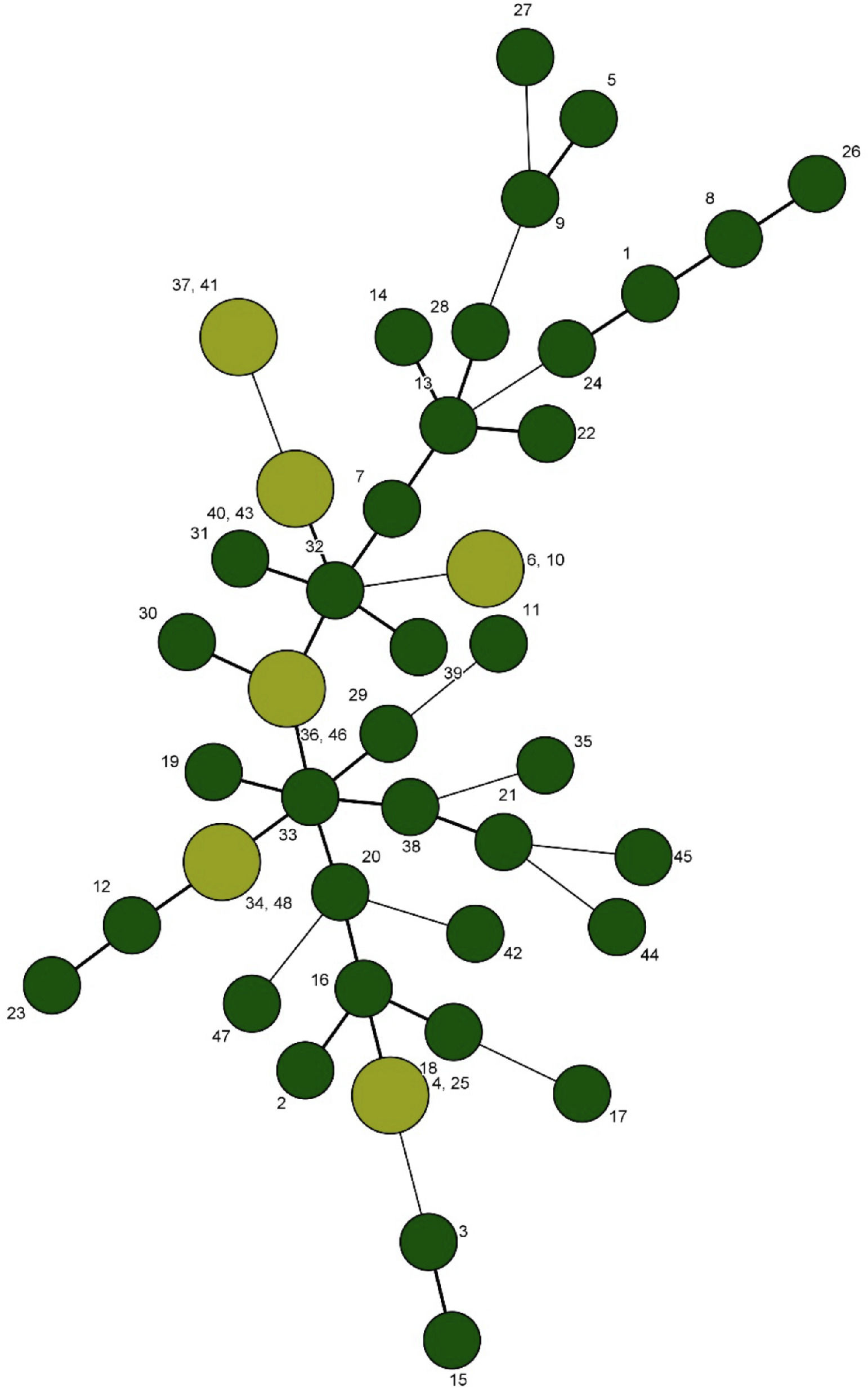
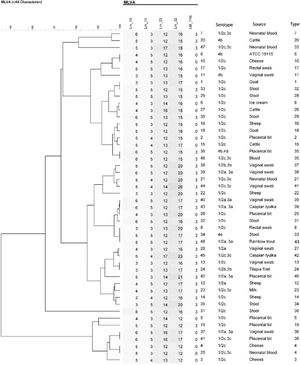
The melting curve and different plots of strain 6 were used as the baseline control. Each strain with equal waveform was grouped. Then, two strains were randomly selected from each group and the PCR reaction was performed for the studied genes using a DNA thermal cycler (PeqLab, Germany) with the following profile: initial denaturation at 94°C for 5 min, 30 cycles of denaturation at 94°C for 30s, annealing at 52°C for 22s, and extension at 72°C for 40s, with a final extension step at 72°C for 5 min. The PCR product was sent for sequencing to Takapozist Company, Iran (on behalf of Bioneer Company, Korea). The allele number was determined and imputed for each locus into BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium) to draw the UPGMA dendrogram and the minimum spanning tree for all the studied L. monocytogenes.
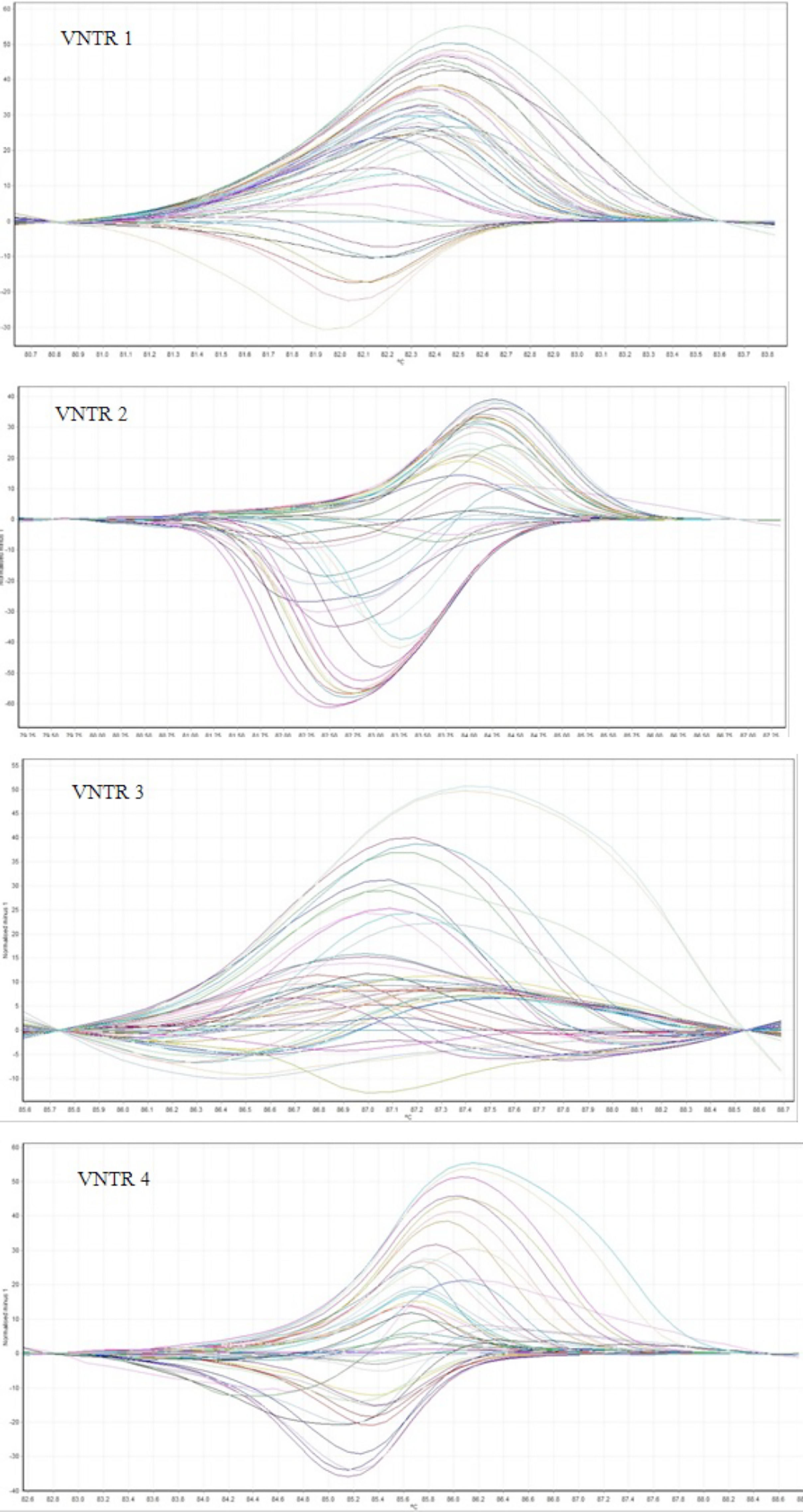
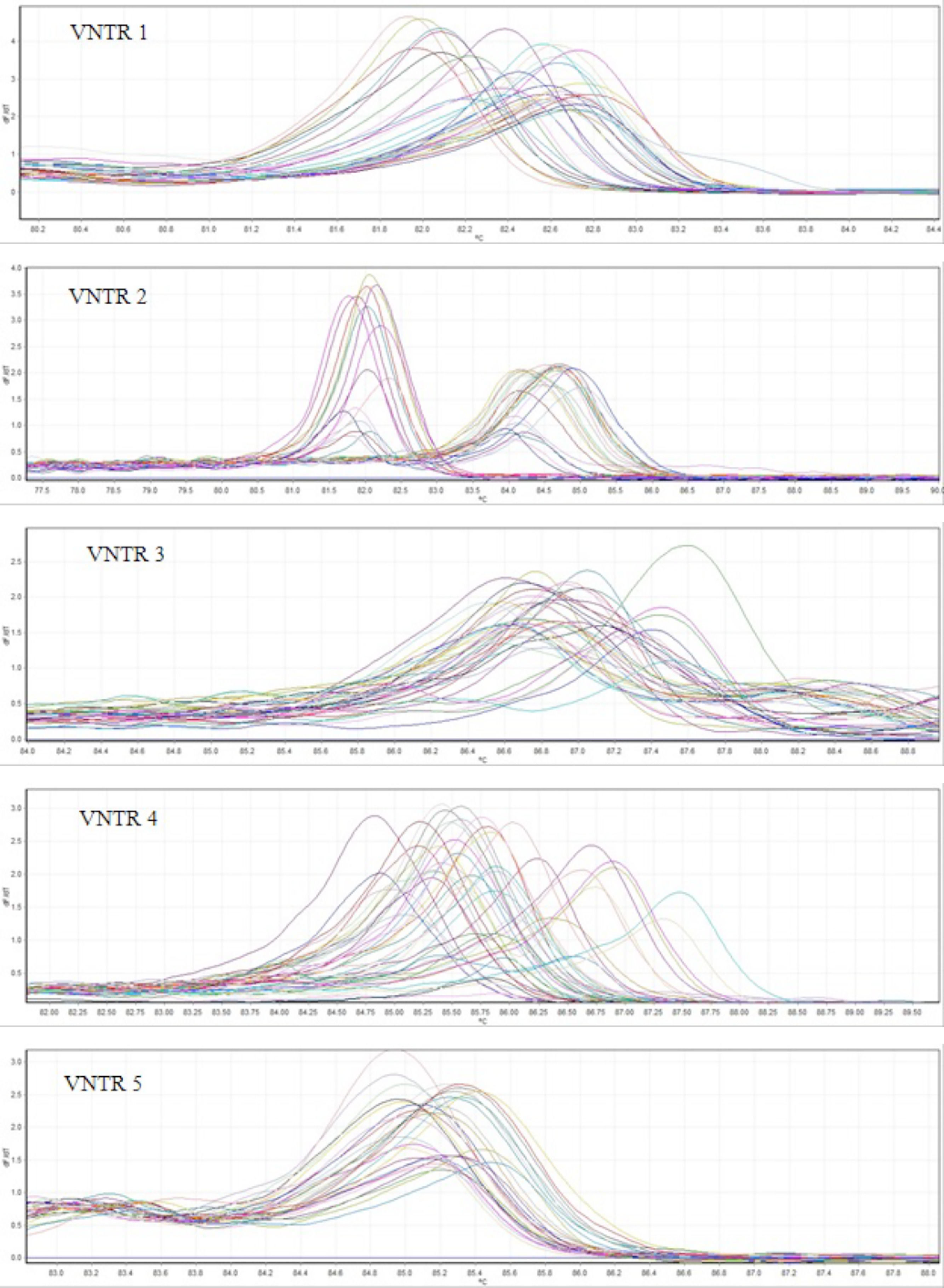
ResultsMLVA-HRMFor the 48 L. monocytogenes strains, we performed HRM on 5 VNTR loci to determine the STs. The different plot and melting curves of 48 L. monocytogenes strains for 5 VNTR regions were drawn (Figs. 1 and 2). HRM was performed in three replications, which gave very similar melting curves, thus confirming the reproducibility of the data. According to similar peak waveforms for 48 L. monocytogenes strains, the Lm_10, Lm_11, Lm_23, Lm_32, and LM_TR6 loci were divided into 6, 3, 5, 7, and 2 types, respectively.
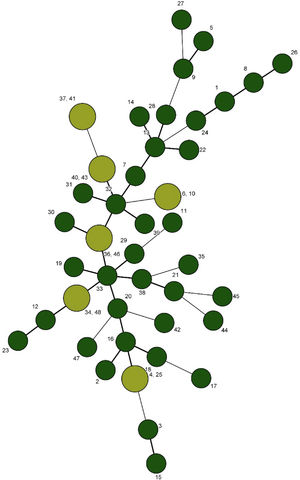
After sequencing and calculating the number of alleles and imputing each locus into BioNumerics and combining the allele numbers of the 5 loci, UPGMA dendrogram and the minimum spanning tree for 48 strains of L. monocytogenes were drawn and 48 strains of L. monocytogenes were finally divided into 43 genotypes. Of these, 38 were single genotypes and each of the 33, 35, 36, 39 and 4 genotypes were determined in 2 isolates. While the genotype 33 in strain originated from vaginal swab and neonatal blood, genotype 4 in strain originated from cheese and neonatal blood, genotype 36 in strain from vaginal swab and placental bit, genotype 35 in strain from placental bit and blood and genotype 39 in strain originated from Caspian tyulka and vaginal swab were observed (Figs. 3 and 4).
Molecular typing of L. monocytogenes isolates plays an important role in identifying the source of infection and preventing the spread of this pathogen.20 Currently, various methods are used for molecular typing of L. monocytogenes from various sources, including PFGE, MLST, and more recently Whole Genome Sequencing (WGS), which requires a lot of work and cost.21,22 MLVA is another molecular typing method that comes with practical benefits such as speed and ease of use. This method uses electrophoresis for diagnosis.23 VNTR loci in the bacterial genome often mutate, leading to changes in the number of tandem repeats and successive changes in nucleic acid. Changes in the number of VNTR repeats are used by the MLVA method for the molecular typing of different isolates.13,24 To evaluate the appropriateness and the suitability of the MLVA method for routine monitoring and subtyping of L. monocytogenes isolates from meat products, Belén Martín et al. collected 113 isolates of L. monocytogenes from meat.25. Their study showed that MLVA is a reliable method for L. monocytogenes typing with a higher discriminating power than MLST, especially for serotype 1/2c isolates.25 Lindstad et al. compared MLVA with the PFGE method to evaluate the resolution power of the MLVA method for molecular typing of L. monocytogenes isolates and their results showed that the MLVA method was slightly more discriminant for Norwegian isolates (28 MLVA profiles and 24 PFGE profiles).26
In HRM, changes in nucleotide sequences and diversity in the chain length of PCR products are indicated by changes in the melting curve.27 In a study by Ohshima et al., analysis of MLVA was performed using the HRM method as a simple and rapid method for differentiating L. monocytogenes isolates. The study also compared the ability of MLVA-HRMA, MLVA using capillary electrophoresis, and MLST to differentiate between strains. This study demonstrated that the MLVA-HRM method was more discriminant than MLST and MLVA using capillary electrophoresis.28 In a study by Lüdeke et al., for phylogenetic analysis, MLST and MLVA were performed on 58 Vibrio parahaemolyticus isolates, and the results gained by both methods were compared with the PFGE patterns obtained in their previous study. Their results showed that HRM-MLVA was able to separate isolates in the same PFGE cluster with the same ST.29
In this study, MLVA-HRM method with five VNTR loci used as a simple and fast typing method was able to differentiate L. monocytogenes strains, which we believe can be a valuable alternative to costly and time-consuming techniques. Also, due to the characteristics of this method, it can be used in routine bacterial diagnoses.
This study was financially supported by Iran University of Medical Sciences (Tehran, Iran), for which we are very grateful.