To investigate the pathogenesis of bloodstream infection by Staphylococcus epidermidis, using the molecular epidemiology, in high-risk neonates.
MethodsWe conducted a prospective study of a cohort of neonates with bloodstream infection using central venous catheters for more than 24h. “National Healthcare Safety Network” surveillance was conducted. Genotyping was performed by DNA fingerprinting and mecA genes and icaAD were detected by multiplex-PCR.
ResultsFrom April 2006 to April 2008, the incidence of bloodstream infection and central venous catheter-associated bloodstream infection was 15.1 and 13.0/1000 catheter days, respectively, with S. epidermidis accounting for 42.9% of episodes. Molecular analysis was used to document the similarity among six isolates of bloodstream infection by S. epidermidis from cases with positive blood and central venous catheter tip cultures. Fifty percent of neonates had bloodstream infection not identified as definite or probable central venous catheter-related bloodstream infection. Only one case was considered as definite central venous catheter-related bloodstream infection and was extraluminally acquired; the remaining were considered probable central venous catheter-related bloodstream infections, with one probable extraluminally and another probable intraluminally acquired bloodstream infection. Additionally, among mecA+ and icaAD+ samples, one clone (A) was predominant (80%). A polyclonal profile was found among sensitive samples that were not carriers of the icaAD gene.
ConclusionsThe majority of infections caused by S. epidermidis in neonates had an unknown origin, although 33.3% appeared to have been acquired intraluminally and extraluminally. We observed a polyclonal profile between sensitive samples and a prevalent clone (A) between resistant samples.
Central venous catheters (CVCs) are widely used in adults and newborns and are the leading cause of bloodstream infection (BSI) in hospitals.1 Infections associated with the use of CVCs represent 10–20% of hospital infections (IHs) in adult patients2 and approximately half of those observed in neonates.3 Infections related to the use of CVC (CR-BSI), i.e. those in which the microorganism detected in the bloodstream is also present in the catheter tip, represent up to 29% of these infections in neonatal intensive care units (NICUs) with an incidence varying from 2 to 49/1000 days CVC.4
The pathogenesis of CR-BSI is multifactorial and complex. Data from studies in adults showed two main potential routes of contamination of the catheter tip with BSI: dissemination of intraluminal microorganisms through contamination of the hub in long-term CVCs, and extraluminally due to migration of microorganisms to the skin at the site of insertion in those short duration CVC.5,6
Although evaluation of samples by molecular techniques is essential, due to their discriminatory power, their use in investigating the pathogenesis of BSIs associated/related to CVC has been scarce; moreover, there are few studies on the pathogenesis of CVC-related BSI in critical newborns.7,8
Gram-negative rods are the main pathogens of neonatal sepsis in developing countries.9 In NICUs, following the adoption of sophisticated tertiary neonatal care with a high rate of invasive device use, coagulase-negative staphylococci (CoNS) stand out as the main agents of neonatal nosocomial sepsis, with Staphylococcus epidermidis being the most frequently isolated from late onset sepsis, accounting for 70% of cases.10,11 Neonatal infection by CoNS is less severe but causes significant morbidity, especially among infants of very low birth weight.12
The main factor in the pathogenicity of S. epidermidis is its ability to form biofilms mediated by polysaccharide intercellular adhesin (PIA), encoded by the ica operon.13 This bacterium adheres to the surface of invasive devices such as CVCs and forms biofilms, which account for its greater resistance to antibiotics, particularly to meticillin, increasing the length of hospital stay and costing around $ 2 billion US dollars just in the United States.14 The bacteria that form biofilms are difficult to eradicate and it is estimated that this mechanism is associated with 65.0% of infections in hospitals.15
ObjectiveTo investigate the pathogenesis of CR-BSI by S. epidermidis in high-risk neonates through a prospective nested cohort study in a randomized trial using molecular epidemiology to determine the origin of microorganisms responsible for BSI.
Materials and methodsPatients, study design and settingThis was a prospective cohort study of patients with a CVC with laboratory confirmed primary BSI by S. epidermidis conducted in the NICU with 10 level-3 beds within a 500-bed teaching hospital from April/2006 to April/2008.
The population analyzed was followed five times/week from birth until discharge or death. Ethical approval was obtained from the Ethics Committee of Uberlândia Federal University according to the requirements of the Ministry of Health.
The study sample consisted of neonates who had at least one CVC placed for longer than 24h, followed-up through epidemiologic vigilance as part of the “National Healthcare Safety Network” (NHSN).
DefinitionsLaboratory-confirmed primary BSIIsolation of recognized pathogens from two blood cultures that were not related to infection at another site, with temperature >38°C and with clinical signs of sepsis, including apnea, temperature instability, lethargy, feeding intolerance, worsening respiratory distress, or hemodynamic instability.16
Catheter tip colonizationAbsence of signs of infection at the catheter insertion site and microorganism growth ≥103CFU/mL at the catheter tips (by quantitative culture) or ≥15UFC/mL (by the “roll- plate” technique).17
CVC-associated BSIBacteraemia (isolation of the same organism with identical antibiogram from blood drawn from peripheral veins and CVC), clinical manifestations of sepsis, defervescence after removal of the implicated catheter, but without laboratory confirmation of CVC colonization.18
Definite catheter-related BSIFor neonates with BSI that did not have an identified source, definite catheter-related BSI was defined by a culture of a peripheral, percutaneously obtained blood sample that was positive for the same organism found to be colonizing the catheter hub or tip (i.e., concordant colonization of the catheter hub or tip).19
Probable catheter-related BSIProbable catheter-related BSI was defined as BSI without an identified source that met one of the following criteria: (1) a blood sample obtained through a peripherally inserted central catheter (PICC) positive on culture for the same organism recovered from the catheter tip or from the catheter hub; culture of a percutaneously obtained peripheral blood sample that either yielded no growth or was not performed; and, if CoNS was isolated, clonal similarity confirmed by restriction-fragment DNA subtyping; or (2) a peripheral blood sample positive for CoNS on culture with similarity between the organism recovered from peripheral blood and that recovered from the catheter tip or from the catheter hub as demonstrated by speciation and antibiogram results, but restriction-fragment DNA subtyping was not done.19
Determination of pathogenesisBSI was classified as extraluminally acquired if there was similarity between the isolate recovered from the catheter tip and isolates recovered from blood samples, either similar or not to isolates recovered from insertion-site samples. BSI was classified as intraluminally acquired if similarity was demonstrated solely between isolates recovered from hub samples and isolates recovered from the blood. If results suggested both routes of acquisition, then the pathogenesis of the BSI was classified as indeterminate.19
Microbiological techniquesHemocultureBlood specimens were obtained from peripheral puncture. Hemocultures were performed by inoculating 1.0mL of blood into a flask of the automatic commercial system Bactec/Alert (Vitek System). Positive cultures were further subcultured onto plates with Blood Agar.
CVC tipCatheters were removed when no longer required for patient care, when the patient experienced an adverse event, or when catheter exchange was deemed necessary. Catheters were removed under aseptic conditions; their tips were cut off with sterile scissors and transferred into sterile tubes and transported to the Microbiology Laboratory. Quantitative culture and “roll-plate” techniques were carried out according to the methods of Brun-Buisson et al.20 and Maki et al.,21 respectively, and were considered positive when ≥103CFU/mL and ≥15CFU/mL were present, respectively.
Skin of CVC insertion siteThe collection of skin material was performed seven and 14 days after catheter insertion, with a sterile saline pre-wet swab over a 20cm2 area. The swab was put into a tube with 1mL sterile saline and then stirred in a vortex; about 0.1mL of the liquid was inoculated onto blood agar plates, which were incubated at 35°C for 24h. Cultures were considered positive if growth of ≥200UFC/20cm2 was seen.22
Nasal mucosa and catheter hubQualitative cultures of material collected from the nostrils and the catheter hub were performed. The swab was put into a tube containing 1mL sterile saline and then stirred in a vortex; about 0.1mL of the liquid was inoculated onto blood agar plates for 24h at 35°C.
Microorganism identificationThe isolates obtained from clinical samples were inoculated onto blood agar and Gram-stained in order to determine their purity and observe their morphology and specific color. After determining these characteristics, the strains were submitted to catalase and coagulase tests.
To identify S. epidermidis, the tests described by Bannerman23 and MAcFaddin24 were used: production of bound coagulase factor (“clumping”) and coagulase-free production of hemolysis, pyrrolidonyl arylamidase enzyme, urease, ornithine decarboxylase, novobiocin susceptibility and fermentation of the carbohydrates mannitol, mannose and trehalose. Molecular analyses were performed on coagulase negative Staphylococcus using the RapID STAPH PLUS system (Oxoid) to confirm their identity.
Molecular techniquesBacterial strainsA total of 12 S. epidermidis strains isolated from blood, catheter tip, and nasal secretion were evaluated. Two reference strains were used: (1) ATCC 35984 S. epidermidis as a positive control for the mecA and icaAD genes; (2) ATCC 12228 S. epidermidis as a negative control for icaAD gene. The strains were stored at −20°C in tryptic soy broth (TSB) (Difco Laboratories, MD, USA) with 20% glycerol.
- (a)
Detection of the mecA and icaAD genes:
To assess the presence of mecA and icaAD genes in S. epidermidis a multiplex-PCR was performed that has previously been described by Santos et al.,25 Araújo et al.26 and McClure et al.27 The primers for the mecA gene detection were: mecA-MRS1 (5′TAGAAATGACTGAACGTCCG3′) and mecA-MRS2 (5′TTGCGATCAATGTTACCGTAG3′), and the primers for the icaAD gene were: icaAD-F (5′TAGTAATCACAGCCAACATCTT3′) and icaAD-R (5′AAACAAACTCATCCATCCGAAT3′). Cycling conditions were as follows: initial denaturation at 94°C for 3min, followed by 30 cycles of denaturation at 94°C for 1min, annealing at 55°C for 1min, and extension at 72°C for 2min, followed by a final extension step at 72°C for 5min. The amplified products were analyzed by electrophoresis on 2.0% agarose gels and stained with ethidium bromide stain.
- (b)
Analysis of the macrorestriction profiles of chromosomal DNA after cleavage with the restriction enzyme SmaI and pulsed field electrophoresis.
Analysis of macrorestriction profiles of chromosomal DNA after cleavage with the SmaI restriction enzyme and pulsed field electrophoresis was performed according to the method described by Nunes et al.28 The gel was processed using the Contour-Clamped Homogeneous Electric Field (CHEF) DR III System (Bio-Rad Laboratory Pty-Ltd.), using a pulse time increasing from 1 to 35s during 6V/cm electrophoresis 23h and 13°C with an angle of 120°. The gel was stained with ethidium bromide (0.5μg/mL) for 30min, destained for 1h with distilled water and observed under ultraviolet light and then photographed.
Differences between isolates were determined by visual inspection of the bands, as recommended by Van Belkum et al.29 and by the Molecular Analyst Fingerprinting Plus software package (version 1.12) of the Image Analysis System (Bio-Rad, Hercules, CA, USA), using the Dice index and the unweighted pair group method with arithmetic averages for estimations of similarity and clustering.
ResultsA total of 63 of 318 neonates from the randomized trial were included in this cohort study of the pathogenesis of CR-BSI in high-risk neonates with CVC who had at least one CVC placed for more than 24h, totalling 5674 patient-days and 4845 catheter-days. There was an average of 1.5 catheters/patient, with 461 episodes of CVC catheterization.
The incidence of laboratory-confirmed primary BSI per 1000 catheter days was 15.1, and 12.6 for neonates weighing ≤1500g. The incidence of CVC-associated to BSI was 13.0 per 1000 CVC days, respectively. The overall mortality rate was 13.5%, and in neonates with low weight (≤1500g) it was 16.1% (Table 1).
Rates of bloodstream infection associated with and related to CVC, catheter tip colonization and mortality in 318 neonates between April 2006 and April 2008.
| Rates | n (%) |
|---|---|
| Laboratory-confirmed primary BSI per 1000 patient-days | 12.9 |
| Laboratory-confirmed primary BSI per 1000 catheter-days | 15.1 |
| Catheter tip colonization per 1000 catheter-days | 17.3 |
| CVC-associated BSI per 1000 catheter-days | 13.0 |
| CVC-related BSI per 1000 catheter-days | 2.1 |
| Overall mortality | 43 (13.5) |
| Mortality of CVC-associated BSI | 30 (69.8) |
| Mortality of CVC-related BSI | 1 (2.3) |
BSI, bloodstream infection; CVC, central venous catheter.
The overall catheter tip colonization rate was 18.2%. S. epidermidis was the microorganism isolated in half (48.7%) of the positive cultures from the CVC tip (Table 2) followed by S. aureus (33.3%). On average, colonized CVCs were in use for 21.9 days. The main agents of microbiologically confirmed primary BSI were S. epidermidis (39.7%), S. aureus (24.6%), and Gram-negative bacilli (12.2%). S. epidermidis was associated with 43.0% of CVC-associated BSI.
Pathogenesis of bloodstream infection by S. epidermidis in high-risk neonates with central venous catheter.
| CVC tip | Blood | Hub | Skin | Nasal mucosa | Type CVC | Technical cultive CVC tip | Length of use (days) |
|---|---|---|---|---|---|---|---|
| S. epidermidis | – | – | – | – | Umbilical | SQ | 2 |
| S. epidermidis | – | – | – | – | Phlebotomy | QT | 3 |
| S. epidermidis | – | – | – | – | Phlebotomy | SQ/QT | 3 |
| S. epidermidis | – | – | – | – | Umbilical | SQ | 4 |
| S. epidermidis | – | – | – | – | Phlebotomy | SQ | 4 |
| S. epidermidis | S. agalactiae | – | – | – | Umbilical | SQ/QT | 4 |
| S. epidermidis | – | – | – | – | Umbilical | SQ/QT | 5 |
| S. epidermidis | – | – | S. epidermidis | – | Umbilical | SQ/QT | 5 |
| S. epidermidis | – | – | S. epidermidis | S. epidermidis | Umbilical | SQ/QT | 5 |
| S. epidermidis | S. aureus | S. aureus | – | S. aureus | PICC | SQ/QT | 5 |
| S. epidermidis | – | – | – | – | PICC | SQ/QT | 6 |
| S. epidermidis | S. aureus/S. haemolyticus | – | S. aureus | S. epidermidis | Umbilical | SQ | 6 |
| S. epidermidis | S. aureus | – | – | – | Umbilical | SQ/QT | 6 |
| *S. epidermidis | *S. epidermidis | – | – | S. epidermidis | Phlebotomy | SQ | 6 |
| S. epidermidis | S. epidermidis | – | – | – | Umbilical | SQ/QT | 6 |
| *S. epidermidis | *S. epidermidis | – | – | – | Flebotomia | SQ | 7 |
| *S. epidermidis | *S. epidermidis | – | – | – | Umbilical | SQ | 8 |
| S. epidermidis | S. bovis | – | S. lugdunensis | – | Umbilical | SQ | 8 |
| S. epidermidis | – | – | – | S. epidermidis | PICC | SQ | 9 |
| S. epidermidis | C. albicans | – | – | – | Phlebotomy | SQ/QT | 9 |
| *S. epidermidis | *S. epidermidis | – | – | – | PICC | SQ | 10 |
| S. epidermidis | – | S. aureus | – | S. aureus | PICC | SQ | 10 |
| *S. epidermidis | *S. epidermidis | – | – | – | Phlebotomy | SQ/QT | 11 |
| S. epidermidis | – | – | – | – | Umbilical | SQ | 12 |
| S. epidermidis | – | – | – | S. aureus | Phlebotomy | SQ/QT | 15 |
| S. epidermidis | – | – | – | – | PICC | SQ | 16 |
| S. epidermidis | S. liquefaciens/E. agglomerans | – | S. epidermidis | – | Phlebotomy | SQ/QT | 17 |
| S. epidermidis | – | – | S. epidermidis | – | PICC | SQ/QT | 19 |
| S. epidermidis | – | S. aureus | S. epidermidis | S. epidermidis | Phlebotomy | SQ/QT | 19 |
| S. epidermidis | – | – | S. epidermidis | – | Phlebotomy | SQ | 20 |
| *S. epidermidis | *S. epidermidis | S. epidermidis | – | *S. epidermidis | PICC | SQ | 23 |
| S. epidermidis | A. baumannii/E. coli | – | – | – | PICC | SQ/QT | 24 |
| S. epidermidis | – | – | – | – | PICC | QT | 25 |
| S. epidermidis | – | – | – | S. aureus | Phlebotomy | SQ | 28 |
| S. epidermidis | – | – | – | – | Phlebotomy | SQ | 32 |
| S. epidermidis | – | – | – | – | Phlebotomy | SQ | 32 |
| S. epidermidis | – | – | S. epidermidis | – | Phlebotomy | SQ | 33 |
| S. epidermidis | – | – | – | – | PICC | SQ | 39 |
SQ, semi-quantitative technique; QT, quantitative technique; CVC, central venous catheter; PICC, peripherally inserted central catheter.
Only one case was considered to be definite CR-BSI, and it was extraluminally acquired. The others were considered probable CR-BSIs, with one probable extraluminally and another probable intraluminally acquired BSI (Table 3).
Results of cultures from 6 cases of BSI, with positive blood and CVC tip cultures, and clonal profiling of the mecA and icaAD genes in samples of S. epidermidis isolated from high-risk neonates.
| Infected patient | Sites | BSI classification | Route of acquisition | CVC type | Catheterization time (days) | PFGE | mecA gene | icaAD gene |
|---|---|---|---|---|---|---|---|---|
| 1 | Blood | Definite | Definite extraluminal | PICC | 10 | A | Positive | Positive |
| CVC tip | CR-BSI | A | Positive | Positive | ||||
| 2 | Blood | Not related | Indeterminate | Umbilical | 8 | H | Negative | Negative |
| CVC tip | C | Negative | Negative | |||||
| 3 | Blood | Not related | Indeterminate | Phlebotomy | 11 | F | Positive | Positive |
| CVC tip | A | Positive | Positive | |||||
| 4 | Blood | Probable | Probable extraluminal | Phlebotomy | 7 | H | Negative | Negative |
| CVC tip | CR-BSI | ND | Negative | Negative | ||||
| 5 | Blood | Not Related | Indeterminate | Phlebotomy | 6 | G | Negative | Negative |
| CVC tip | A | Positive | Positive | |||||
| Nasal mucosa | D | Negative | Negative | |||||
| 6 | Blood | Probable | Probable intraluminal | PICC | 23 | E | Positive | Negative |
| CVC tip | CR-BSI | B | Negative | Negative | ||||
| Hub | ND | Positive | Positive |
ND, not done; BSI, bloodstream infection; PFGE, pulsed field gel electrophoresis; PICC, peripherally inserted central catheter.
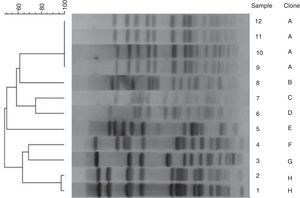
Results from DNA typing of S. epidermidis isolates by PFGE are summarized in Fig. 1. No temporal relationship was observed. Macrorestriction analyses of DNA singled out the presence of one prevalent clone in the unit (clone A). Among 12 isolates analyzed, 8 PFGE patterns were observed. PFGE type A was positive for the mecA and icaAD genes. In only one patient the same clone (A) was detected in blood and on the CVC tip. Different clone profiles of isolates were observed in colonized and infected patients, and in CVC of some patients. Among sensitive samples that did not carry the icaAD gene, we observed a polyclonal profile without predominant types. Clone A was related to the same samples recovered from blood and CVC tip of the patient (Table 4).
Dendogram of genotypic strains of Staphylococcus epidermidis after fragmentation with restriction enzyme SmaI and computer analysis of PFGE.
Samples 11 and 12: blood and CVC tip, respectively, from patient 1. Samples 2 and 7: blood and CVC tip, respectively, from patient 2. Samples 4 and 9: blood and CVC tip, respectively, from patient 3. Sample 1: blood sample from patient 4. Samples: 3, 6 and 10: blood, nasal mucosa and CVC tip, respectively, from patient 5. Samples: 8 and 5: blood sample and CVC tip, respectively, from patient 6.
Strain relatedness of 12 isolates categorized according to PFGE type and genotype by PCR in 6 cases of BSI due to S. epidermidis.
| PCR profile | Genotype | PFGE | PFGE patterns | Predominant clone (%) | Same patient | |
|---|---|---|---|---|---|---|
| Blood | CVC tip | |||||
| All | 12 | |||||
| 1 | mecA+/icaAD+ | 5 | A, F | A (80.0) | Yes | Yes |
| 2 | mecA−/icaAD− | 6 | H, C, G, D, B | H (33.3) | No | No |
| 3 | mecA+/icaAD− | 1 | E | E (100.0) | No | No |
PFGE, pulsed field gel electrophoresis.
Most strains of S. epidermidis (66.7%) isolated from blood and catheter tips were resistant to oxacillin, with the mecA gene detected in 50.0% of the samples. Approximately 67.0% of the samples obtained from blood and 50.0% of those from CVC tips had the ability to form biofilms, and the icaAD gene was detected in two and three of six samples cultured from blood and catheter tip, respectively. Multiplex PCR for the mecA and icaAD genes was performed for 14 S. epidermidis resistant to oxacilin, and 50% and 42.9% of the isolates presented an amplicon consistent with the mecA and icaAD genes, respectively. All isolates showed resistance to a 1mg/mL oxacilin disk diffusion test and were considered resistant by agar screening with oxacilin (data not shown).
DiscussionOrganisms responsible for CR-BSI can access a catheter either extraluminally or intraluminally, from the skin at the insertion site of the catheter or by hematogenous spread of organisms from a distant focus of infection and contamination of the catheter hub or, in the case of an outbreak, via contaminated intravenous solution, respectively. According to the literature, in adults, extraluminal acquisition appears to be the most frequently found in non-cuffed catheters and non-tunneled short term CVC.5,6
Unlike the study conducted by Garland et al.19 in which the intraluminal pathway was the main source of contamination of CVCs (67%), in our study with neonates, considering the genotypic analysis, we demonstrated that most BSI was indetermined and only one was from a confirmed extraluminal source. This failure to confirm the contamination routes of CVC strongly suggests significant differences in the pathogenesis of CR-BSI in neonates and adults.
In the present study, high-risk neonates with BSI caused by S. epidermidis had a CVC inserted for an average of 12.3 days and we demonstrated that most CVC insertions were via long-term phlebotomy (39.5%). The high percentage of phlebotomy use in this series and the high percentage of contamination of the mucosa (40%) in the neonates could explain the results. Based on this study, our neonatal unit reduced the use of phlebotomy considerably and increased the use of PICC.
Catheter colonization rate was lower in our study than the rate reported by Garland et al.,19 in which 22% of 106 cultures obtained grew an organism. In our study the catheter hub was found to be also colonized in only 4 (10.5%) of 38 hub cultures when a CVC tip was colonized. Differences in study design and our sterile saline moistened swab of the hub may explain some of the discrepancies in colonization rates between these two studies.
Another important finding that reinforces the problems of handling catheters was the subsequent detection of colonization by S. aureus contaminating 75% of CVCs hubs analyzed. This aspect was also reported by Mahieu et al.30 who showed an association between handling the hub and an increased risk of CR-BSI and nosocomial sepsis in neonates. The reduction in the incidence of BSI in our NICU was gradual and significant, and appeared to occur prominently over two specific periods, namely from 2006 to 2008 and from 2010 to 2012. During the second period we adopted standard procedures for insertion of PICC and continuous care. This strategy was based on our findings as well as other studies conducted in the unit.31 Although improved aseptic procedures and careful insertion of CVCs were documented to reduce the incidence of CR-BSI rates in our unit, these procedures were not adhered to resulting again in an increase in infection rates, thus reinforcing the importance of measures to control and prevent infection at the unit.
S. epidermidis was responsible for 43% of all CA-BSIs. While S. epidermidis has long been considered non-pathogenic, it is now recognized as a relevant opportunistic pathogen in hospitals.32 Phenotypic methods demonstrated that 66.7% and 50% of blood samples and CVC tips, respectively, were producing biofilms, with detection of the icaAD gene in 33.3% of the blood samples and 50% of CVC tips. The presence of the mecA gene was also common among these isolates (50%).
Based on our findings about the pathogenesis CR-BSI the pathogenesis of these infections seem to be different than what has been reported for adults.
ConclusionsIn conclusion, there seems to be evidence that pathogenesis of CR-BSI in neonates differs from that reported in adults, as the majority of these infections that occurred in high risk neonates presented routes of indeterminate acquisition and only 16.7% were acquired extraluminally. Findings of this study reinforce the need for combined strategies to prevent extraluminally and intraluminally acquired BSI in high-risk neonates in developing countries.
Conflicts of interestThe authors declare no conflicts of interest.