Dengue, the most prevalent arboviral disease worldwide, is caused by any of the four dengue virus (DENV) serotypes that co-circulate constantly in hyperendemic areas such as Medellin (Colombia), and these serotypes are transmitted by mosquitoes of the genus Aedes. In this study, we evaluated the replicative capacity of strains isolated in Medellin between 2003 and 2007 in C6/36 cells and in colonies of Aedes aegypti collected during 2010–2011 from high or low-incidence areas within the same city. The phylogenetic analysis grouped isolates according to the predominant genotypes found in the Americas, and the in vitro characterization showed differences in the morphological changes induced by the isolates of each of the isolated serotypes compared to the reference serotypes. In vitro replicative capacity studies demonstrated that genomic copy number increased at four days post-infection and that cell viability decreased significantly compared to the control for all serotypes. The largest number of genomic copies in C6/36 was produced by DENV-2, followed by DENV-1 and DENV-4; DENV-3 produced the smallest number of genomic copies and had the smallest negative effect on cell viability. Finally, differences in the in vivo replication of intercolonial serotypes between the Rockefeller colony and the field colonies and among the intracolonial serotypes were found. The replication of DENV-2 at 7 and 14 days in both high- and low-incidence colonies was higher than that of the other serotypes, and replication of DENV-3 in the mosquito colonies was the most stable on the days evaluated. Our results support the notion that replication and, possibly, DENV transmission and severity depend on many factors, including serotype and vector characteristics.
Dengue, a disease that is endemic in many countries, is classified as the main viral disease transmitted by arthropods (arbovirosis). Although it has been reported that 50–200 million cases of dengue occur annually, 1 recent studies have suggested that this number is underestimated and that the most likely number of cases per year is approximately 400 million, of which symptomatic cases represent only 96 million, with the remainder of cases being asymptomatic.2 In addition, the World Health Organization has estimated that in the past five decades the number of cases of dengue has increased 30-fold. Thus, it is urgent that existing control strategies be implemented and improved to achieve a significant reduction in the number of cases in the coming years.3 These cases are distributed in at least 125 countries that are located in tropical and subtropical regions, with Southeast Asia and the Western Pacific being the regions with the highest incidence, followed by the region of the Americas (primarily Latin America).1
The etiological agent of this disease is the dengue virus (DENV), of which four serotypes (DENV-1 to DENV-4) belonging to the urban viral cycle have been identified.4 This virus belongs to the family Flaviviridae and the genus Flavivirus and has a positive-sense RNA genome5 that encodes a single polyprotein that is cleaved by proteases of cellular and viral origin, resulting in the production of three structural proteins (C, M and E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3A, NS4B, NS4B and NS5).6 Based on the variability of their genomes, specific genotypes have been identified within each serotype. Five DENV-1, six DENV-2, five DENV-3, and five DENV-4 genotypes are currently known.7
Colombia, a country located in the northern part of South America, is one of the Latin American countries most affected by dengue. The first epidemics of dengue in Colombia were reported in the 1950s with the presence of DENV-1, DENV-2, and DENV-4 serotypes; these serotypes have spread throughout the country during the following decades. Their re-emergence occurred in the early nineteenth century with the introduction of DENV-3. In Colombia, the disease is classified as endemic and even hyperendemic in some cities, presenting up to 150,000 cases in certain years (such as in 2010).8
The DENV is transmitted by the bite of infected mosquitoes of the genus Aedes (Diptera: Culicidae), with Aedes aegypti being the most important species in urban areas. Although the vector is mainly distributed in regions located below 1800 m.a.s.l., in Colombia it has been detected in regions above that altitude.9 Since the re-emergence of serotype 3 in Colombia in 2002, the four serotypes of the DENV have been constantly co-circulating. Approximately 80% of the cases reported between 2009 and 2010 were associated with serotypes 1 and 2, and the remaining 20% were associated with serotypes 3 and 4.8 Although there is insufficient evidence to determine the association between serotype and disease severity in Colombia, only serotype 3 has not been associated with cases of severe dengue.8 In addition to the problem of endemicity, the recent emergence of Aedes albopictus, a species that is considered a more competent vector for DENV infection,10 in urban areas as well as the recent description of different lineages of A. aegypti,11 which is considered the most efficient species in the transmission of DENV, aggravate the problem in the country.
The dynamics of dengue transmission involve a complex set of factors related to the virus. These factors include its vertebrate and invertebrate hosts, which under certain spatio-temporal conditions allow the vector to transmit the virus.12 Several approaches, both in vitro (using mammalian or mosquito cell lines) and in vivo (using mosquito colonies), have been used to evaluate factors that may positively or negatively affect viral transmission; one of these factors is viral replication.
In vitro, the C6/36 cell line, which was derived from A. albopictus larvae, is widely used to study the virus-cell interaction. This cell line is highly susceptible to infection by all four serotypes and is easy to grow and maintain.13 Differences have been reported in the ability of viral serotypes to enter and replicate in C6/36 cells. For example, two polypeptides of molecular weights 80 and 67 kDa14 and two glycoproteins of 40 and 45 kDa15 that are present in the membranes of these cells are used by several strains of DENV-2 for their entry and subsequent replication. In contrast, several strains of DENV-3 and DENV-4 use only one laminin-binding protein of 37–67 kDa molecular weight for this process.16 These results indicate that although the line is highly susceptible to infection, there are differences in the viral replication process dependent on the infectious serotype.
In vivo, colonies of A. aegypti adapted to laboratory conditions, such as the Rockefeller colony (ROCK).17 have been used to study the replication and the transmission capacity of DENV in the vector. This transmission capacity is related to extrinsic or ecological factors, such as climatic conditions, the abundance of vertebrate hosts and vector population densities, as well as intrinsic factors, such as the genetic basis of the vector, which is also known as vector competence.18,19 In turn, replication of the virus in a given vector is used as an indirect measure of the ability of that vector to transmit the virus. For example, it has been reported that genetic variability among mosquito populations is an important factor in susceptibility to infection and competence for DENV transmission, with differences in susceptibility to infection of mosquito populations from different geographical regions20 and among laboratory populations of A. aegypti compared to field populations.21 These facts show that the differences between A. aegypti populations from different countries or continents or even within the same country may be associated with differences in the DENV vector competence of these mosquitoes. Conversely, it has also been demonstrated that the vector competence of populations of African A. aegypti for both DENV and yellow fever virus (another virus transmitted by A. aegypti) is dependent on the infecting viral genotype and on the population of the infected vector.22
Considering that some of the four DENV serotypes circulating in Colombia may have a greater potential for transmission by the vector and that there is substantial genetic variation among mosquito populations,11,23 another factor that could be related to the transmission capacity of the virus, our objective was to evaluate the replicative capacity of four Colombian clinical isolates in the C6/36 cell line and in colonies of A. aegypti collected in areas of high and low incidence of the disease in the city of Medellin (Colombia) as an experimental model to evaluate the capacity of viral transmission.
MethodologyMaintenance of the cell line and production of clinical isolates and reference virusesThe cell line derived from A. albopictus C6/36HT (HT – high temperature, clone growth at 34 °C) was provided by Dr. María Guadalupe Guzmán of the Pedro Kourí Institute (Havana, Cuba). This cell line was maintained in DMEM supplemented with 10% fetal bovine serum. The clinical isolates (DENV-1/MedCol, DENV-2/MedCol, DENV-3/MedCol and DENV-4/MedCol) obtained during 2003–2007 were donated by the Laboratorio Departamental de Salud de Antioquia [Departmental Laboratory of Public Health of Antioquia].24 These isolates and the reference strains DENV-1 West Pac 74, DENV-2 S16803, DENV-3 CH53489, DENV-4 TVP360, which were obtained from the National Institute for Biological Standards and Control, were amplified in C6/36HT cells. At 11 days post-infection, the supernatants were harvested and stored at −70 °C for in vitro and in vivo infection assays.
Selection of the collection area of the entomological materialBased on information provided by the authorities responsible for epidemiological control in the city of Medellin (Colombia), the areas of the city with the highest and lowest reported incidences of dengue cases during the first half of 2010 were selected. To ensure an adequate number of artificial stagnant water reservoirs capable of harboring a sufficient number of mosquitoes in their aquatic stages, areas that housed populations of low socioeconomic strata were selected because residents of these areas frequently store rain water. Likewise, to guarantee good population densities of the vector, areas with Breteau Indexes between 5 and 50 were selected, indicating a moderate risk of A. aegypti Infestation in such areas. Thus, the Las Violetas neighborhood was selected as the high-incidence area (H-INC), and the Progreso 2 and El Triunfo neighborhoods were selected as the low-incidence area (L-INC). It should be noted that neighborhoods had to be selected to form the L-INC area so that the sizes of both areas (H-INC and L-INC) were comparable, thus avoiding sampling bias (Fig. 1).
Collection of entomological materialThe first part of the sampling was conducted on November 3, 4 and 5, 2010, in the H-INC area. In this area, 79 homes were inspected, and entomological material was collected (86 larvae, 23 pupae and 7 adults) in 11 of them. The second part of the sampling was conducted on February 14, 15, 16 and 17, 2011, in the L-INC area. In this area, 83 homes were inspected, and entomological material was collected (149 larvae and 25 pupae) in 12 of them. All of the entomological material collected was taken to the insectarium for breeding and maintenance.
Establishment of field colonies and taxonomic verification of speciesColonies referred to as H-INC or L-INC area field colonies were established from the entomological material collected in each area. Both field colonies (H-INC, L-INC) and the colony used as a control (ROCK) were maintained at an average temperature of 28 ± 2 °C, 75 ± 5% relative humidity and a 12-h light/dark photoperiod. The larvae were kept in plastic trays with clean water and fed once daily with sterile Truchina®. The pupae were transferred to breeding cages; when the adults emerged, they were fed a 10% sugar solution ad libitum. For the maintenance of the colonies, mature females were provided with mice as a blood source to promote oviposition and the eggs derived from these females (identified by molecular analysis, as explained later) were collected on paper for positioning and stored in containers that provided optimal conditions for their proper conservation and subsequent use. Once the colonies were established, taxonomic verification was performed with females using previously reported taxonomic keys.25 Experiments were conducted with the endorsement of the Ethics Committee for Animal Experimentation at the University of Antioquia.
Evaluation of viral replication capacity in C6/36HT cellsTwenty-four hours prior to infection, 1.25 × 106 cells/well were plated in 96-well plates. The cells were infected with 1 × 105 genomic copies/mL of each of the viral serotypes. The cell culture supernatants were harvested at 2, 4, 6 and 8 days post-infection and stored at -80 °C for subsequent quantification of viral genomic copies. Monolayers were used to determine cell viability.
Evaluation of viral replication capacity in A. aegypti coloniesFemale mosquitoes with an average age of eight days from each of the three colonies were kept in an insectarium under controlled conditions (temperature 28 ± 2 °C, relative humidity 75 ± 5% and a 12-h light/dark photoperiod). Prior to each of the tests, 80 to 100 individuals from each colony were selected from separate breeding cages from which the sugar solution had been removed 24 h prior to the oral challenge to increase avidity at the time of artificial feeding. For feeds, a viral inoculum (1 × 105 genomic copies) of each of the serotypes was mixed with human blood from a DENV-negative donor (evaluated by ELISA) in a tube containing sodium citrate as an anticoagulant. The mosquitoes were fed using an artificial membrane feeding system (LAUDA, reference A103) for 30 min. Successfully fed females were selected by hand aspiration and placed in a cage, where they were kept for up to 21 days and fed with a 10% sugar solution until infection detection by conventional PCR, immunofluorescence or RT-PCR, depending on the case).
Evaluation of the viability of infected cellsCell viability in cultures of infected C6/36HT cells was evaluated by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) method. In this method, cell monolayers that had been infected for different time periods were incubated with 100 μl MTT (0.5 mg/ml) for 3 h at 37 °C in a 5% CO2 atmosphere protected from light. After the incubation period, the formazan salts were dissolved in dimethyl sulfoxide for 15 min. Finally, the absorbance at 450 nm was read in a Thermo Scientific VarioScan Flash reader.
Quantification of viral replication by plaque formationFor viral quantification, the plating titration technique was used as previously described. Briefly, 5 × 104 VERO cells were plated in 24-well plates, and serial dilutions (1 × 10−1 to 1 × 10−5) of the previously collected viral supernatants were added to the cell monolayers. At two hours after infection, the supernatants were removed, 1.5% carboxymethylcellulose prepared in DMEM supplemented with 2% FBS was added, and the plate was incubated at 37 °C in a 5% CO2 atmosphere for seven days. The cells were then fixed in 4% paraformaldehyde prepared in PBS and stained with 1% crystal violet. The plates were counted to determine the number of plaque-forming units per milliliter (PFU/mL). Because the clinical isolates did not form plaques, the real-time RT-PCR quantification technique was used for viral quantification in both cells and mosquitoes.
Extraction, retrotranscription and conventional PCRFor the extraction of RNA from supernatants and mosquitoes, the modified RNAzol extraction technique was used. For the extraction of RNA from cell cultures, RNA was obtained from 200 μL of supernatant, and for the extraction of RNA from mosquitoes, RNA was obtained from individuals mixed in pools of five mosquitoes. The samples were mixed with 750 μL of RNAzol extraction solution. Subsequently, 200 μL of chloroform was added, and the mixture was homogenized for 20 min on ice. The samples were then centrifuged, and 650 μL of the aqueous phase was mixed with 650 μl of isopropanol to precipitate the RNA overnight at −20 °C. The precipitated RNA was washed with cold 80% ethanol and resuspended in 60 μl of RNAse-free water for quantification in Nanodrop® (Thermo Scientific). For the retrotranscription process, 0.5 μg of RNA, 0.5 μg/mL of random primers (Promega), 0.5 mM dNTPs and 200 U of M-MLV retrotranscriptase (Promega) were used. The reaction was incubated at 37 °C for 1 h, and the cDNA was stored at −20 °C until use. From 2 μL of cDNA, a fragment of the C-prM region of each of the four DENV serotypes was amplified using the primers and the thermal profile previously reported.26 The PCR products were verified by electrophoresis on 1.5% agarose gels. To verify the integrity of the RNA obtained from mosquitoes, conventional PCR was performed for the detection of constitutive expression of the β-actin gene.
Phylogenetic analysis of viral isolatesFor the identification of the genotypes, the E gene (1479–1485 nucleotides) of each strain was amplified and sequenced using the Sanger system. The partial sequencing products were assembled and edited using the DNASTAR program (Lasergene, Madison, WI, USA). These sequences were aligned with 19 to 21 sequences obtained from GenBank per serotype and selected to represent all known genotypes of each serotype (2–5 sequences per genotype). The alignment was performed with the Muscle program. A phylogenetic analysis was then performed by the maximum parsimony (MP) system using the subtree-pruning-regrafting (SPR) algorithm. The reliability of the tree was measured by performing 100 bootstrap replicates. These analyses were performed in the MEGA 7.0 package.27
Viral quantification by RT-qPCRThe amplification of each of the four serotypes from individual mosquitoes (n: 8 for any experimental condition) or infected cell cultures was performed according to a previously reported protocol.26 Briefly, the first step of DNA polymerase activation (95 °C for 10 min) was performed, followed by 35 cycles of denaturation at 95 °C for 15 s, annealing at 50 °C for 15 s and extension at 72 °C for 20 s. After the final extension, a dissociation curve was performed to confirm the specificity of the products. The number of genomic copies was calculated by absolute quantification using standard curves that were constructed and have been previously reported.28
Statistical analysisThe statistical analysis was conducted using SPSS for Windows (version 15.0). The Shapiro–Wilk test was used to determine compliance with the normality assumption of the data obtained from the cells, and the Kolmogorov–Smirnov test was used for the data obtained from the mosquitoes. Subsequently, the Mann–Whitney U test was used to identify significant differences between two independent groups that were not normally distributed. In all cases, a probability of p < 0.05 in a two-tailed test was considered statistically significant.
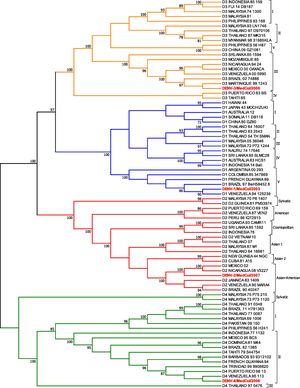
ResultsPhylogenetic analysisPhylogenetic analysis by maximum parsimony showed that the DENV-1/MedCol strain belongs to subtype V of serotype 1, the DENV-2/MedCol strain belongs to the Asian-American genotype of serotype 2, the DEVN-3/MedCol strain belongs to subtype III of serotype 3, and the DENV-4/MedCol strain belongs to subtype II of serotype 4. These genotypes have predominated in the Americas in recent decades (Fig. 2).
Phylogenetic analysis of clinical isolates. Cladogram resulting from the maximum parsimony analysis of the E gene sequences of the DENV strains used in this study (highlighted in red) and 77 other sequences that together are representative of all genotypes of each serotype. The branches corresponding to serotypes DENV-1, DENV-2, DENV-3 and DENV-4 are shown in blue, red, orange and green, respectively. The branch support obtained by "bootstrap" is shown close to the corresponding node. The analysis was performed using the MEGA 7.0 program.
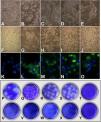
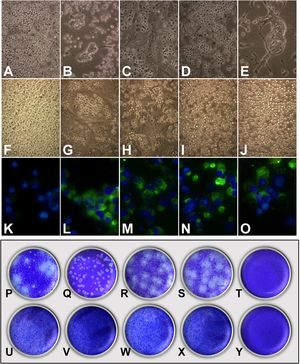
Infections of C6/36HT cells were performed with the clinical isolates of each of the four serotypes and with the reference viruses. At 11 days post-infection, cytopathic effects were observed; these were more evident in cultures infected with the reference strains (Fig. 3B–E) than in those infected with the clinical isolates (Fig. 3G–J). The DENV-3/MedCol strain (Fig. 3I) and the DENV-4/MedCol strain (Fig. 3J) induced the fewest morphological changes compared to the uninfected control (Fig. 3F). Infection by the isolates was confirmed with the identification of viral antigen by immunofluorescence using serotype-specific monoclonal antibodies (Fig. 3K–O). Plaque formation capacity in semisolid medium was evaluated for each of the clinical isolates, but none of these isolates induced the formation of plaques (Fig. 3U–X), unlike the reference strains (Fig. 3P–S), for which plaque formation was evident. Therefore, quantification to determine the number of viral genomic copies resulting from infection with the clinical isolates was conducted by qPCR.
Infection and quantification of C6/36HT cells. Upper panel: Cells were infected with viral isolates and reference viruses; at 11 days post-infection, the cytopathic effect was documented (A–J), and the presence of viral antigen was confirmed by direct immunofluorescence (K–O). A, F and K: Control cells without infection. B: DENV-1/West Pac 74. C: DENV-2/S16803. D: DENV-3/CH53489. E: DENV-4/TVP360. G and L: DENV-1/MedCol. H and M: DENV-2/MedCol. I and N: DENV-3/MedCol. J and O: DENV-4/MedCol. Lower panel: Titration of the virus by plating of culture supernatants from infected C6/36HT cells on semisolid medium. P: DENV-1/West Pac 74. Q: DENV-2/S16803. R: DENV-3/CH53489. S: DENV-4/TVP360. U: DENV-1/MedCol. V: DENV-2/MedCol. W: DENV-3/MedCol. X: DENV-4/MedCol. T-Y: Control without infection. Only the reference viruses were able to form plaques in semisolid medium.
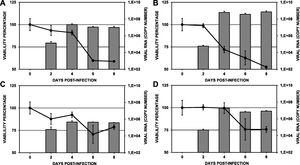
The viability of infected C6/36HT cells was evaluated by the MTT method every 48 h until 8 days post-infection. In general, as the duration of infection increased, cell viability decreased (Fig. 4). In the cells infected with the DENV-1/MedCol strain, statistically significant differences in viability compared to day 0 were found from day 4 onward (p < 0.05, viability percentages of 91.0%, 59.7% and 58.9% on days 4, 6 and 8 post-infection, respectively). Similarly, cells infected with the DENV-2/MedCol strain showed statistically significant differences in viability compared to day 0 from day 4 onward (p < 0.05, viability percentages of 72.1%, 62.7% and 52.4% on days 4, 6 and 8 post-infection, respectively). In the cultures infected with the DENV-3/MedCol and DENV-4/MedCol strains, statistically significant differences in viability were observed only on day 8 (p < 0.05, viability percentages of 78.7% and 75.5%, respectively, compared to day 0). The percentage viability of cells infected with each of the serotypes over time is shown in Table 1.
Viability and replication of clinical isolates in C6/36HT cell cultures. Cultures were maintained for eight days; cell viability was measured by the MTT method and quantification of infection by RT-qPCR was performed every 48 h. A. DENV-1/MedCol. B: DENV-2/MedCol. C: DENV-3/MedCol. D: DENV-4/MedCol. A comparison of the viability of cells infected by each of the serotypes over time is shown in Table 1, and the error bars represent the standard error of the mean. Comparison of the replication of the different clinical viral isolates at various times after infection is shown in Table 2. Viability decreased significantly from day 4 in the cultures infected with the DENV-1/MedCol and DENV-2/MedCol strains but only until day 8 in the cultures infected with the DENV-3/MedCol and DENV-4/MedCol strains. The highest replication occurred at day 4 in the DENV-2/MedCol strain. Grey Bars: Viral Replicaction. Continuous line: Viability percentage. The error bars represent the standard error of the mean (n:4).
Effect of viral infection on cell viability.
| Viability of different serotypes of dengue virus | |||||
|---|---|---|---|---|---|
| DAY 0 | DAY 2 | DAY 4 | DAY 6 | DAY 8 | |
| DENV-1 | 100 | 93.4 | 91.0 | 59.7 | 58.9 |
| p = 0.37 | p = 0.03 | p = 0.00 | p = 0.00 | ||
| DENV-2 | 100 | 98.8 | 72.1 | 62.7 | 52.4 |
| p = 0.65 | p = 0.00 | p = 0.00 | p = 0.00 | ||
| DENV-3 | 100 | 87.8 | 92.4 | 70.6 | 78.7 |
| p = 0.18 | p = 0.09 | p = 0.20 | p = 0.01 | ||
| DENV-4 | 100 | 100.4 | 99.5 | 75.7 | 75.7 |
| p = 0.91 | p = 0.95 | p = 0.29 | p = 0.01 | ||
When evaluating the genomic copy number of each of the serotypes in C6/36HT cells, we found that the peak replication for all serotypes occurred at four days post-infection; the DENV-1/MedCol, DENV-2/MedCol, DENV-3/MedCol and DENV-4/MedCol strains showed increases in viral copy number of 160.3-, 1037610376.8-, 7.9- and 412.8-fold, respectively, relative to day 2. The highest replication titer was found in cultures infected with DENV-2/MedCol (6.05 × 108 genomic copies), followed by cultures infected with DENV-1/MedCol or DENV-4/MedCol (2.16 × 107 and 1.9 × 107 genomic copies, respectively). The lowest replication occurred in cultures infected with DENV-3/MedCol (4.64 × 105 genomic copies) (Fig. 4). When comparing the replication titers of each of the serotypes at each time point tested, statistically significant differences (p < 0.05) were found only on day 6; at that time, the replication of the DENV-2/MedCol strain (3.65 × 108 genomic copies) was greater than that of the DENV-4/MedCol strain (6.61 × 106 genomic copies), and the replication of the latter was greater than that of the DENV-3/MedCol strain (4.39 × 105 genomic copies). On day 8, the replication of the viral strains followed the order DENV-2 > DENV-1 > DENV-4 > DENV-3; the numbers of genomic copies of each of these strains present on day 8 were 7.11 × 108, 9.85 × 106, 8.33 × 106, and 3.72 × 105, respectively. A comparison of the replication of the different clinical viral isolates on each day is shown in Table 2.
Comparison of in vitro viral replication among the different serotypes on each of the evaluated days.
| Viral replication in vitro of different serotypes of dengue virus | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DAY 2 | DAY 4 | DAY 6 | DAY 8 | ||||||||||||
| Vs. | DENV-2 | DENV-3 | DENV-4 | Vs. | DENV-2 | DENV-3 | DENV-4 | Vs. | DENV-2 | DENV-3 | DENV-4 | Vs. | DENV-2 | DENV-3 | DENV-4 |
| DENV-1 | 5.8 × 104 | 5.9 × 104 | 4.7 × 104 | DENV-1 | 6.1 × 108 | 4.6 × 105 | 1.9 × 107 | DENV-1 | 3.7 × 108 | 4.4 × 105 | 6.6 × 106 | DENV-1 | 7.1 × 108 | 3.7 × 105 | 8.3 × 106 |
| 1.4 × 105 | p = 0.67 | p = 0.67 | p = 0.33 | 2.2 × 107 | p = 0.33 | p = 0.33 | p = 1.00 | 1.1 × 107 | p = 0.06 | p = 0.07 | p = 0.11 | 9.9 × 106 | p = 0.03 | p = 0.11 | p = 0.34 |
| DENV-2 | – | 5.9 × 104 | 4.7 × 104 | DENV-2 | – | 4.6 × 105 | 1.9 × 107 | DENV-2 | – | 4.4 × 105 | 6.6 × 106 | DENV-2 | – | 3.7 × 105 | 8.3 × 106 |
| 5.8 × 104 | – | p = 1.00 | p = 0.67 | 6.1 × 108 | – | p = 0.33 | p = 0.33 | 3.7 × 108 | – | p = 0.06 | p = 0.03 | 7.1 × 108 | – | p = 0.06 | p = 0.06 |
| DENV-3 | – | – | 4.7 × 104 | DENV-3 | – | – | 1.9 × 107 | DENV-3 | – | – | 6.6 × 106 | DENV-3 | – | – | 8.3 × 106 |
| 5.9 × 104 | – | – | p = 1.00 | 4.6 × 105 | – | – | p = 0.33 | 4.4 × 105 | – | – | p = 0.03 | 3.7 × 105 | – | – | p = 0.01 |
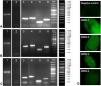
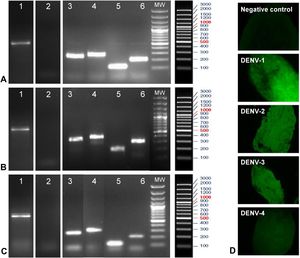
Before beginning the evaluation of DENV viral replication in A. aegypti mosquitoes from the H-INC and L-INC colonies, it was necessary to rule out the possibility that these mosquito populations were naturally infected with any of the four serotypes of the virus. Lane 1 in panels A, B and C of Fig. 5 shows the mRNA encoding the β-actin protein, which was used as a marker of RNA integrity in the mosquito samples, obtained from the ROCK colony (Panel A), the H-INC colony (Panel B) and the L-INC colony (Panel C); lane 2 in each of the three panels shows the detection of natural infection. It was determined that none of the three colonies were naturally infected with any of the four DENV serotypes.
Evaluation of natural infection and susceptibility to experimental infection in colonies of A. aegypti. Panels A–C: Detection of natural infection and susceptibility to experimental infection. Panel A. ROCK Colony. Panel B: L-INC Colony. Panel C: H-INC Colony. Panel D: Confirmation of experimental infection by immunofluorescence. Representative photomicrographs of viral antigen detection in the midgut of mosquitoes of the ROCK control strain are shown. Lanes 1: Amplification of b-actin. Lanes 2: Detection of natural Infection by PCR-Multiplex. Lanes 3–6: Adult females artificially fed each of the four DENV serotypes were sacrificed after 14 days and processed for detection of DENV by conventional PCR. Lanes 3, detection of DENV-1 (260 bp); lanes 4, detection of DENV-2 (288 bp); lanes 5, detection of DENV-3 (119 bp); and lanes 6, detection of DENV-4 (208 bp). MW: molecular weight markers.
Detection of the viral genome after experimental infection of the mosquito colonies with each of the four viral isolates is shown in lanes 3–6 of panels AC in Fig. 5. In lane 3 of each panel, a band of 260 bp corresponding to the DENV-1/MedCol strain can be observed; in lane 4 of each panel, a 288-bp band corresponding to the DENV-2/MedCol strain can be observed; in lane 5 of each panel, a 119-bp band corresponding to the DENV-3/MedCol strain is observed; and in lane 6 of each panel, a 208-bp band corresponding to the DENV-4/MedCol strain is observed. The susceptibility of the mosquito colonies to infection by each of the four clinical isolates was confirmed by immunofluorescence; representative photomicrographs of such detection in the midgut of ROCK colony mosquitoes are shown in Panel D of Fig. 5.
Comparison of the viral replication of different serotypes of A. aegypti in different mosquito coloniesIn general terms, replication of the DENV-1/MedCol strain remained constant between day 7 (Fig. 6A) and day 14 (Fig. 6B) in the three mosquito colonies, with a slight decrease on day 21 (Fig. 6C) in the ROCK colony. In the case of the DENV-2/MedCol strain, replication in the ROCK colony remained stable between day 7 and day 14 (Fig. 6A and B), but it decreased markedly (by three logarithmic units) in the H-INC and L-INC colonies. The replication of the DENV-3/MedCol strain was the most stable in all three colonies at the three-time points evaluated. Finally, replication of the DENV-4/MedCol strain increased at day 14 (Fig. 6B) in the ROCK and H-INC colonies and decreased in the L-INC colony (Fig. 6).
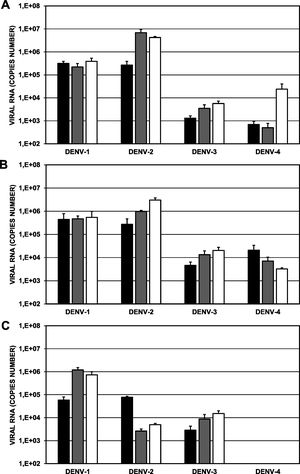
Replication of clinical isolates in different populations of A. aegypti. Approximately 100 female mosquitoes were artificially fed a blood/viral inoculum mixture (1 × 105 genomic copies) and were maintained for up to 21 days after feeding. Viral detection was performed by RT-qPCR in individual mosquitoes. A, Day 7; B, Day 14; and C, Day 21. Black bars, ROCK colony; gray bars, H-INC colony; white bars, L-INC colony. Comparisons of intra- and intercolonial replication are presented in Tables 3 and 4, respectively. The error bars represent the standard error of the mean (n:8).
Comparison of the replication of each of the four strains in the ROCK colony versus that in the H-INC and L-INC colonies showed that at day 7 (Fig. 6A), only the replication of the DENV-2/MedCol strain (4.24 × 106 genomic copies) was significantly higher (p < 0.05) in the L-INC colony than in the ROCK colony (2.67 × 105 genomic copies); at day 14 (Fig. 6B), the DENV-2/MedCol strain was found in greater copy numbers in both the L-INC colony (3.03 × 106 genomic copies) and in the H-INC colony (9.78 × 105 genomic copies) than in the ROCK colony (2.75 × 105 genomic copies, p < 0.05). Finally, at day 21 (Fig. 6C), it was found that the replication of the DENV-1/MedCol strain was higher both in the H-INC colony (1.20 × 106 genomic copies) and in the L-INC colony (7.31 × 105 genomic copies) than in the ROCK colony (2.7 × 105 genomic copies, p < 0.05); higher replication of the DENV-3/MedCol strain in the L-INC colony (1.52 × 104 genomic copies) than in the ROCK colony (2.88 × 103 genomic copies, p < 0.05) was also observed. Finally, the replication of the DENV-2/MedCol strain was lower in both the L-INC colony (4.93 × 103 genomic copies) and the H-INC colony (2.63 × 103 genomic copies) than in the ROCK colony (7.69 × 104, p < 0.05). The comparison of the replication of the different viral isolates in different colonies is shown in Table 3.
Viral replication among intercolonies of A. aegypti of different serotypes of Dengue virus.
| Viral replication among intercolonies of A. aegypti | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| DAY 7 | DAY 14 | DAY 21 | |||||||
| ROCK | H-INC | L-INC | ROCK | H-INC | L-INC | ROCK | H-INC | L-INC | |
| DENV-1 | 3.2 × 105 | 2.2 × 105 | 3.9 × 105 | 4.7 × 105 | 4.7 × 105 | 5.4 × 105 | 2.7 × 105 | 1.2 × 106 | 7.3 × 105 |
| p = 0.46 | p = 0.66 | p = 0.95 | p = 0.87 | p = 0.00 | p = 0.02 | ||||
| p = 0.35 | p = 0.88 | p = 0.27 | |||||||
| DENV-2 | 2.7 × 105 | 6.0 × 106 | 4.2 × 106 | 2.8 × 105 | 9.7 × 105 | 3.0 × 106 | 7.7 × 104 | 2.6 × 103 | 4.9 × 103 |
| p = 0.07 | p = 0.00 | p = 0.03 | p = 0.03 | p = 0.00 | p = 0.00 | ||||
| p = 0.40 | p = 0.06 | p = 0.06 | |||||||
| DENV-3 | 1.3 × 103 | 3.5 × 103 | 5.7 × 103 | 4.7 × 103 | 1.3 × 104 | 2.0 × 104 | 2.9 × 103 | 8.8 × 103 | 1.5 × 104 |
| p = 0.23 | p = 0.06 | p = 0.20 | p = 0.10 | p = 0.36 | p = 0.03 | ||||
| p = 0.35 | p = 0.48 | p = 0.37 | |||||||
| DENV-4 | 6.9 × 102 | 5.0 × 102 | 2.4 × 104 | 2.1 × 104 | 7.1 × 103 | 3.2 × 103 | No data | No data | No data |
| p = 0.66 | p = 0.22 | p = 0.47 | p = 0.40 | ||||||
| p = 0.22 | p = 0.34 | ||||||||
Finally, when comparing replication between serotypes within the same mosquito colony, we observed the following. In the ROCK colony at day 7 (Fig. 6A, black bar), replication of the DENV-1/MedCol strain (3.2 × 105 genomic copies; p < 0.05) was greater than that of the DENV-3/MedCol (1.3 × 103 genomic copies) and DENV-4 (6.9 × 102 genomic copies) strains, and at day 21 (Fig. 6C, black bar), replication of both the DENV-1/MedCol strain (2.7 × 105 genomic copies) and the DENV-2/MedCol strain (7.7 × 104 genomic copies) was greater than that of the DENV-3/MedCol strain (2.9 × 103 genomic copies, p < 0.05). In the H-INC colony, significant differences were found only at days 14 and 21. At day 14 (Fig. 6B, gray bar), replication of the DENV-2/MedCol strain (9.9 × 105 genomic copies) was greater than that of the DENV-1/MedCol (4.7 × 105 genomic copies), DENV-3/MedCol (1.3 × 104 genomic copies) and DENV-4/MedCol (7.1 × 103 genomic copies) strains, and replication of the DENV-1/MedCol strain was greater than that of the DENV-3/MedCol (1.3 × 104 genomic copies) and DENV-4/MedCol (7.1 × 103 genomic copies) strains. At day 21 (Fig. 6C, gray bar), replication of the DENV-1/MedCol strain (1.2 × 106 genomic copies) was greater than that of the DENV-2/MedCol (2.6 × 103 genomic copies) and DENV-3/MedCol (8.8 × 103 genomic copies) strains. Finally, in the L-INC colony at day 7 (Fig. 6A, white bar), replication of the DENV-2/MedCol strain (4.2 × 106 genomic copies) was higher than that of the DENV-1/MedCol (3.9 × 105 genomic copies), DENV-3/MedCol (5.7 × 103 genomic copies) and DENV-4/MedCol (2.4 × 104 genomic copies) strains, and replication of the DENV-4/MedCol strain (2.4 × 104 genomic copies) was greater than that of the DENV-3/MedCol strain (5.7 × 103 genomic copies). At day 14 (Fig. 6C, white bar), replication of the DENV-2/MedCol strain (3.0 × 106 genomic copies) was higher than that of the DENV-1/MedCol (5.4 × 105 genomic copies), DENV-3/MedCol (2.0 × 104 genomic copies) and DENV-4/MedCol (3.0 × 103 genomic copies) strains. At day 21 (Fig. 6C, white bar), only the replication of the DENV-1/MedCol strain (7.3 × 105 genomic copies) was greater than that of the DENV-2/MedCol (4.9 × 103 genomic copies) and DENV-3/MedCol (1.5 × 104 genomic copies) strains. The comparison of the replication of the different viral isolates in the different mosquito colonies is shown in Table 4.
Intercolonial viral replication among of A. aegypti of different serotypes of Dengue virus.
| Intracolonial viral replication among of A. aegypti | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DAY 7 | DAY 14 | DAY 21 | |||||||||
| Vs. | DENV-2 | DENV-3 | DENV-4 | Vs. | DENV-2 | DENV-3 | DENV-4 | Vs. | DENV-2 | DENV-3 | DENV-4 |
| Rockefeller colony | |||||||||||
| DENV-1 | 2.7 × 105 | 1.3 × 103 | 6.9 × 102 | DENV-1 | 2.8 × 105 | 4.7 × 103 | 2.1 × 104 | DENV-1 | 7.7 × 104 | 2.9 × 103 | No data |
| 3.2 × 105 | p = 0.75 | p = 0.00 | p = 0.00 | 4.7 × 105 | p = 0.67 | p = 0.24 | p = 0.25 | 2.7 × 105 | p = 0.31 | p = 0.02 | |
| DENV-2 | – | 1.3 × 103 | 6.9 × 102 | DENV-2 | – | 4.7 × 103 | 2.1 × 104 | DENV-2 | – | 2.9 × 103 | No data |
| 2.7 × 105 | p = 0.16 | p = 0.16 | 2.8 × 105 | p = 0.26 | p = 0.28 | 7.7 × 104 | p = 0.00 | ||||
| DENV-3 | – | – | 6.9 × 102 | DENV-3 | – | – | 2.1 × 104 | DENV-3 | – | – | No data |
| 1.3 × 103 | p = 0.18 | 4.7 × 103 | p = 0.42 | 2.9 × 103 | |||||||
| High incidence colony | |||||||||||
| DENV-1 | 6.9 × 106 | 3.5 × 103 | 5.0 × 102 | DENV-1 | 9.9 × 105 | 1.3 × 104 | 7.1 × 103 | DENV-1 | 2.6 × 103 | 8.8 × 103 | No data |
| 2.2 × 105 | p = 0.07 | p = 0.08 | p = 0.08 | 4.7 × 105 | p = 0.02 | p = 0.02 | p = 0.02 | 1.2 × 106 | p = 0.00 | p = 0.00 | |
| DENV-2 | – | 3.5 × 103 | 5.0 × 102 | DENV-2 | – | 1.3 × 104 | 7.1 × 103 | DENV-2 | – | 8.8 × 103 | No data |
| 6.9 × 106 | p = 0.07 | p = 0.07 | 9.9 × 105 | p = 0.00 | p = 0.00 | 2.6 × 103 | p = 0.30 | ||||
| DENV-3 | – | – | 5.0 × 102 | DENV-3 | – | – | 7.1 × 103 | DENV-3 | – | – | No data |
| 3.5 × 103 | p = 0.13 | 1.3 × 104 | p = 0.38 | 8.8 × 103 | |||||||
| Low incidence colony | |||||||||||
| DENV-1 | 4.2 × 106 | 5.7 × 103 | 2.4 × 104 | DENV-1 | 3.0 × 106 | 2.0 × 104 | 3.0 × 103 | DENV-1 | 4.9 × 103 | 1.5 × 104 | No data |
| 3.9 × 105 | p = 0.00 | p = 0.05 | p = 0.06 | 5.4 × 105 | p = 0.03 | p = 0.29 | p = 0.27 | 7.3 × 105 | p = 0.02 | p = 0.02 | |
| DENV-2 | – | 5.7 × 103 | 2.4 × 104 | DENV-2 | – | 2.0 × 104 | 3.0 × 103 | DENV-2 | – | 1.5 × 104 | No data |
| 4.2 × 106 | p = 0.00 | p = 0.00 | 3.0 × 106 | p = 0.02 | p = 0.02 | 4.9 × 103 | p = 0.05 | ||||
| DENV-3 | – | – | 2.4 × 104 | DENV-3 | – | – | 3.0 × 103 | DENV-3 | – | – | No data |
| 5.7 × 103 | p = 0.00 | 2.0 × 104 | p = 0.08 | 1.5 × 104 | |||||||
Dengue virus replication in both mammalian cells and mosquito cells has been extensively documented, and viral replication has been reported in A. aegypti as an indirect measure of vector competence. However, these studies have several drawbacks because they are almost always performed with only serotype 229–31 or with reference viruses.32,33 Furthermore, laboratory-adapted mosquito colonies, which have been shown to behave differently with respect to infection than field colonies,34 are typically used in vector competence evaluations. For this reason, we evaluated the replication of four clinical isolates obtained in the city of Medellin, Colombia (DENV-1/MedCol, DENV-2/MedCol, DENV-3/MedCol, DENV-4/MedCol) in C6/36 cells as well as in reference mosquito colonies (ROCK) and in field colonies (L-INC and H-INC) collected within the same geographical area.
In vitro, the DENV-2/MedCol strain displayed the best replication, with an increase of four logarithmic units relative to day 2 (Fig. 4B). This increase in replication correlated with a strong decrease in viability of the infected cells (ranging from 30% at day 4 to more than 50% at day 8) (Table 1) and a marked cytopathic effect (Fig. 2H). These results are consistent with previous reports showing that at five days post-infection the cytopathic effect in cultures of C6/36 cells infected with DENV-2 increased and that mortality increased by 20% relative to the uninfected control.35 These results are very interesting, considering that serotype 2 has been responsible for the largest number of epidemics worldwide, including Colombia,36 and that serotype 2 virus is also the most associated with disease severity, which is directly related to the replication capacity of the virus.37,38 By contrast, the DENV-3/MedCol strain induced the least pronounced morphological changes in the cultured cells (Fig. 3I), and this observation was correlated with a decreased effect of the virus on cell viability (Fig. 4C). After infection with this strain, viability decreased by only 20% during the entire infection, and less viral replication was observed; of the four serotypes, this was the only one in which viral copy number increased by only one logarithmic unit compared to the uninfected control (7.9 times at day 4). The lower replication of serotype 3 compared to that of serotype 2 under the same experimental conditions has been reported previously for both individual infections and co-infections in mosquito cells lines and laboratory colony.39 In addition, low replicative capacity of this serotype could be correlated with its lack of association with the presence of severe dengue in humans,40 but further studies in mammalian cells could help to validate this findings.
The strains belonging to serotypes 1 and 4 (DENV-1/MedCol and DENV-4/MedCol) yielded very similar results with respect to both cell viability (causing a decrease in viability of not more than 40%, with the DENV-4/MedCol strain having the least effect (Table 1) and viral replication. In the case of replication, we observed an increase of two logarithmic units relative to day 2 of infection, with the increase of the DENV-1/MedCol strain being greatest at day 8, and that of the DENV-4/MedCol strain being greatest at day 4. It is difficult to discuss these results in light of the published literature because these two serotypes are the least studied. Interestingly, on certain days (days 6 and 8) the replication of the DENV-4/MedCol strain was found to be greater than that of the DENV-3/MedCol strain; this observation is consistent with previous reports that titers of serotype 4 in C6/36 cells were up to 1000 times lower than those of DENV-3 at eight days post-infection.13
The observed phenotypic differences in replication capacity, cell viability and the presence or absence of a marked cytopathic effect may be related not only to the infectious serotype of the virus but also to the presence of specific receptors for certain serotypes present on the cell membrane. For example, a laminin-binding protein has been shown to facilitate the entry of DENV-3 into mosquito cells,16 and the HSP70 heat-shock protein facilitates the entry of DENV-2,41 thus the differential entry of the virus depending on the presence of specific receptors would favor the replication of some serotypes. The clinical origin of the samples could be another factor for such replication differences. In that sense, in vivo studies have shown that strains from clinical isolates have greater replication ability in immunocompetent mice than strains adapted to the laboratory.42 However, it is important to note that the strains used in the referenced study were passaged three to five times in C6/36 cells to increase titer and isolated during periods of outbreak,42 which could contribute to increasing their replication capacity in vitro. In our study, the inocula were prepared from clinical isolates with two successive passages in C6/36 to maintain their characteristics as close as possible to those of the virus under field conditions.
The results of the in vivo tests conducted in the present study showed that there are both intercolonial differences (demonstrated by comparing the colonies obtained in the field with the ROCK colony) and intracolonial differences (demonstrated by comparing the replication of each of the serotypes within the same colony) among the strains tested in this study. There were no significant intercolonial differences in the replication of any of the serotypes at any of the times at which the two field colonies (H-INC vs L-INC) were compared, but differences were indeed found among the field strains relative to the ROCK colony (Table 3). The lack of differences between the H-INC and L-INC colonies may be explained by the fact that although these colonies are derived from mosquitoes obtained in areas with very different incidence levels of dengue, there are no differences in their development (larval development or larval survival) or in their phenotype (such as wing size).43 Although we did not perform an identification of the genetic stock of field A. aegypti used in the present study, based on previous reports, it can be assumed that these populations are genetically homogeneous due to anthropogenic factors that have favored allelic homogeneity between the mosquito populations of the two study areas.44 In addition, studies of A. aegypti collected in 2010 from the city of Medellin showed that although genetic differences at the nucleotide level are found in mosquitoes collected in different areas, these mosquito populations all express a prevalent haplotype.23 Another study conducted with populations from Medellin showed that there is low genetic differentiation in the populations of the vector,11 which suggests that although the populations were collected in different areas, phenotypic or genotypic differences that could affect susceptibility to infection, such as midgut scape or infection barriers could be less probable in those colonies. However, mapping of quantitative trait loci in the A. aegypti populations is needed to validate this differences.
In some cases, there were statistically significant differences between the ROCK colony and the field colonies (Table 3). For example, the DENV-2/MedCol strain replicated better both in the H-INC and L-INC colony than in the ROCK colony at days 7 and 14, but its replication was significantly decreased at day 21. Similar behavior was observed in the replication of the DENV-1/MedCol and DENV-3/MedCol strains, which was higher in the H-INC and L-INC colonies than in the ROCK colony at day 21 (Table 3). In this regard, it is important to emphasize that although the origin of the ROCK colony is controversial, it is assumed to be a strain native to Cuba,17 and differences in the replication of DENV-2 in colonies of mosquitoes of different geographic origin have been previously reported.45 Similarly, these differences could be even more marked than indicated in previous studies, considering that phylogeographic analyses have shown that mosquito populations in cities such as Medellin may have been derived from and may be more related to populations of western Africa,11 which may in turn be more susceptible to serotype 2 infection than populations such as ROCK that originated in other geographic regions. Although our study did not focus on the differences in vector competence when evaluating different genotypes of the same viral serotype, it is important to note that both the nature of the virus as well as its serotype and different strains produce variants that result in totally different interactions between the virus and its vertebrate host.22 Hence, it would be interesting to conduct studies of this type that consider the effects of different genotypes belonging to the same serotype.
The largest number of statistically significant differences between the replication of the serotypes was found in the intracolonial analysis (Table 4). In the ROCK colony, we see that at day 7 the replication of the DENV-1/MedCol strain is greater than that of the DENV-3/MedCol and DENV-4/MedCol strains, and at day 21 the replication of the DENV-1/MedCol and DENV-2/MedCol strains is greater than that of the DENV-3/MedCol strain. In the H-INC colony, differences in the replication of several serotypes were found beginning on day 14. At day 14, the replication of the DENV-2/MedCol strain was greater than that of the other three strains, and at day 21, the replication of the DENV-1/MedCol strain was greater than that of the DENV-2/MedCol and DENV-3/MedCol strains. In the L-INC colony, several differences were found starting at day 7. In this colony, at both day 7 and day 14, the replication of the DENV-2/MedCol strain was greater than that of the other three serotypes; in turn, the replication of the DENV-4/MedCol strain at day 7 was greater than that of the DENV-3/MedCol strain. Finally, at day 21, replication of the DENV-1/MedCol strain was greater than the replication of the DENV-2/MedCol and DENV-3/MedCol strains. These differences in replication among serotypes within the same mosquito colony are consistent with some previous reports. For example, replication of DENV-2 and DENV-3 is much higher than that of DENV-1 and DENV-4 in mosquito populations in Cabo Verde46 and in Senegal mosquito populations the replication of DENV-3 is lower than that of DENV-1.47 Although the factors responsible for these differences in replication among serotypes have not been fully elucidated, it has been proposed that the differences may be related to natural barriers in the mosquito itself. Thus, for example, the midgut infection barrier may inhibit the early stages of viral replication (adsorption, denudation, transcription-translation), whereas the midgut escape barrier may affect the steps of viral assembly and maturation.48 In a similar way, differences in the innate immune response were found among laboratory and field-derived strains of A. aegypti, which could induced a differential response to dengue virus serotypes.49
Although our in vitro and in vivo results are consistent, the vector competence of the main vector species of the genus Aedes was compared, and it was shown that A. aegypti exhibits a differential response to infection by and dissemination of two different strains of DENV-2 whereas A. albopictus does not show significant differences in its vector competence for the strains evaluated.50 As previously stated, these differences may be due to differences in both the mosquito populations and the viral strains, both of which differ among geographic areas.
ConclusionStrains of the four DENV serotypes isolated in Medellin showed differences in replicative capacity in the C6/36 cell line, and their effects on cell viability were inversely proportional to increases in the number of viral genomic copies at the time points examined. The four DENV serotypes also caused differential cytopathic effects in the cultures, suggesting a relationship between the serotype and the severity of infection. However, it was also demonstrated that viral replication in the vector is affected by the serotype, origin of the colony, and the processes of extrinsic replication. These results suggest that the effectiveness of A. aegypti in transmitting DENV depends on many factors; for this reason, other factors related to viral replication should be studied. However, our findings make a significant contribution to the characterization of the virus-vector relationship and provide a greater understanding of the dynamics of transmission and circulation of the different serotypes of the virus. In the same sense, these results can help prioritize strategies of control and prevention in countries such as Colombia in which DENV is endemic.
FundingThis study was funded by COLCIENCIAS (Departamento Administrativo de Ciencia,Tecnología e Innovación). Project 111549326083. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of interestThe authors declare no conflicts of interest.