At present, there is no standardized marker that is routinely used in clinical laboratories to diagnose coccidioidomycosis. Thus, the goals of this study were to obtain a sequence characterized amplified region (SCAR) marker for the identification of Coccidioides spp., evaluate its specificity and sensitivity in fungal DNA-spiked blood and sputum samples, and compare it with previously described molecular markers. Specific amplified fragment length polymorphism (AFLP) amplicons for Coccidioides immitis and Coccidioides posadasii were cloned into the vector pGEM® -T Easy vector and sequenced to develop a SCAR marker. Oligonucleotides were designed to identify Coccidioides spp. by polymerase chain reaction (PCR), and the specificity and sensitivity of these oligonucleotides were tested with the DNA from related pathogens. The specificity and sensitivity of the SCAR marker was evaluated with blood and sputum samples spiked with Coccidioides DNA and compared with other previously described markers (621, GAC2, and Ag2/PRA). In addition, the conditions for its use were established using biological samples. A specific marker named SCAR300 was obtained to identify Coccidioides spp. that exhibited good sensitivity and specificity. The results showed that all of the markers tested in this study can identify Coccidioides spp. However, the SCAR300 and 621 markers were the most sensitive, whereas the SCAR300 marker was the most specific. Thus, the SCAR300 marker is a useful tool to identify Coccidioides spp.
The fungi Coccidioides immitis and Coccidioides posadasii are the causative agents of coccidioidomycosis,1 a predominant disease in the Americas. The areas most affected by this mycosis are endemic areas of the Southwestern United States of America (USA),2 northern Mexico, some Central America regions3 and South America.4,5 Although most infections caused by these fungi are asymptomatic, some can develop into symptomatic clinical infections with mild respiratory symptoms or may develop into severe disseminated infections that are typically associated with Aids or HIV-infected patients, individuals having received transplants, hemodialysis patients, cancer patients undergoing treatment (primarily Hodgkin’s lymphoma), pregnant women, or individuals with diabetes or tuberculosis.6,7
The number of coccidioidomycosis cases in the USA has increased in recent years.8 However, the number of cases in Mexico is unknown, as coccidioidomycosis ceased to be a notifiable disease in 1994, although it has been suggested that the same trend is occurring.9
The diagnosis of this disease has traditionally been based on the results of a combination of clinical data, the isolation of the causative agent in clinical samples, and imaging studies. However, since the causative fungi grow slowly, rapid methods for its identification are required, such as conventional serological techniques, although these methods also have limitations.10 For this reason, molecular techniques that use different polymerase chain reaction (PCR) markers have been developed in recent years.11–18 However, these techniques have not yet had a significant impact on the majority of clinical laboratories in the diagnosis of coccidioidomycosis. Thus, other identification strategies have been developed, one of which is the use of the so-called SCAR (sequence characterized amplified region) markers, which have been useful for studying variation among organisms, identifying strains of interest, determining the origin of isolates, studying population structure, and detecting pest resistance genes.19 SCAR markers have also been used for diagnostic and epidemiological application in other human pathogenic fungi, such as Histoplasma capsulatum,20 demonstrating its usefulness for the identification of specific microorganisms. Therefore, the aim of this study was to obtain a SCAR marker from polymorphic patterns obtained from amplified fragment length polymorphism (AFLP) to identify Coccidioides spp., in addition to evaluate its specificity and sensitivity in fungal DNA-spiked blood and sputum samples. Furthermore, we compared the SCAR marker to three previously described molecular markers and established the conditions for its use in biological samples.
Material and methodsIsolatesA total of 40 isolates were included in this study (Table S1), including four isolates from Mexico that were previously identified as C. immitis and 36 identified as C. posadasii, of which 25 were isolated in Mexico and 11 in Argentina.21 All of the isolates were cultured in tubes containing Mycobiotic agar® (BD Bioxon, Estado de México, México) and were incubated at 28 °C for five days or until good growth was observed for subsequent trials.
Biological samplesWhole blood and sputum samples used in this study were obtained from a healthy human volunteer.
DNA extraction from C. immitis and C. posadasii isolatesAll of the C. immitis and C. posadasii isolates included in the study were grown in YPD medium (10% yeast extract, 10% peptone, and 20% dextrose) for 7–10 days at 28 °C. DNA extraction was performed as described by Duarte-Escalante et al.21 The DNA concentration was determined via spectrophotometry (Spectrophotometer DS-11, DeNovix, Delawere, USA), and the DNA samples were stored at 4 °C until use.
The DNA from Sporohtrix schenckii was kindly provided by Conchita Toriello (Facultad de Medicina, UNAM, Mexico), Candida glabrata, was kindly provided by María Guadalupe Frías De León (Hospital Regional de Alta Especialidad de Ixtapaluca, México), Histoplasma capsulatum was kindly provided by Maria Lucia Taylor (Facultad de Medicina, UNAM, Mexico), Aspergillus fumigatus and A. niger were kindly provided by María del Rocío Reyes Montes (Facultad de Medicina, UNAM, Mexico), and Mycobacterium tuberculosis was kindly provided by Miriam Bobadilla del Valle (Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico). DNA from all of the fungi tested, as well as from M. tuberculosis, was used to check the specificity of the Coccidioides spp. molecular markers studied.
Obtaining of the SCAR marker: AFLP and SCAR marker selectionThe AFLP assays were performed according to Duarte-Escalante et al.,21 using six selective primer combinations: E + AA:M + CAC, E + AA:M + CAT, E + AA:M + CTG, E + AA:M + CTC, E + AC:M + CAT and E + AC:M + CTC. The analysis of the polymorphic patterns obtained through AFLP with the six combinations of oligonucleotides allowed the identification of a band common in all isolates, of 300 bp with the combination E + AC/M + CAT. The specific DNA fragment of Coccidioides spp. were purified using a QIAquick gel extraction kit (Qiagen, Inc., Valencia, California, USA) and cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA), according to Frías De León et al.20 The SCAR marker was sequenced at the Unidad de Biología Molecular, Instituto de Fisiología Celular, UNAM, using an ABI Prism 3100 automated DNA sequencer (Applied Biosystems, Inc., Foster City, CA, USA). The SCAR marker sequences were analyzed using BLAST22 to verify similarities between all of the fungal sequences deposited in the database. A specific sequence of the fungus which had no coincidence whatsoever with the sequences of related fungi deposited in the GenBank was selected to design the specific nucleotides for Coccidioides spp. Oligonucleotides were designed based on the SCAR sequence using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and were synthesized by Sigma-Genosys (The Woodlands, Texas, USA). The PCR with the SCAR marker conditions were established with the 40 DNA preparations obtained from C. immitis and C. posadasii.
Evaluation of the sensitivity and specificity of the SCAR, Ag2/PRA, and microsatellite 621 and GAC2 markersThe sensitivity of the SCAR marker and that of the Ag2/PRA and microsatellite 621 and GAC2 markers was determined using different concentrations of DNA from the C. posadasii reference strain (HU-1).
The specificities of the SCAR, Ag2/PRA,11 GAC2,1 and 6211 markers were evaluated using DNA from the C. posadasii reference strain (HU-1) and that of other pathogens that cause clinical symptoms similar to Coccidioides spp. (A. niger, A. fumigatus, H. capsulatum, S. schenckii, C. glabrata, and M. tuberculosis).
Evaluation of the SCAR, Ag2/PRA, and microsatellite GAC2 and 621 markers in blood and sputum samples spiked with C. posadasii DNA (HU-1)Five hundred microliters of blood or sputum was spiked with 30 µL of C. posadasii DNA (HU-1) at different concentrations (2.83 × 102, 2.83 × 101, 2.83 × 100, 2.83 × 10−1, 2.83 × 10−2, 2.83 × 10−3, 2.83 × 10−4, 2.83 × 10−5, and 2.83 × 10−6 ng/µL). Each tube was processed to obtain total DNA using a DNeasy Blood & Tissue Kit (Qiagen).
All PCR assays were performed in a total volume of 50 µL with 5, 10, 15, and 20 µL of total DNA obtained from blood or sputum samples spiked with C. posadasii DNA (HU-1). The PCR conditions used for each marker were the same as those described above.
ResultsObtaining of the SCAR marker: selection of AFLP bands for the SCAR markerAnalysis of the polymorphic patterns obtained by AFLP with the six combinations of selective oligonucleotides used resulted in the identification of differential bands between C. posadasii and C. immitis. Four differential bands were obtained for C. posadasii: a 250-bp band obtained with the E + AA/M + CTC combination; two bands, a 150-bp band and a 300-bp band, obtained with the E + AC/M+CAT combination; and a 180-bp band obtained with the E + AA/M + CAT combination, as well as a 200-bp differential band obtained with the E + AA/M + CAT combination. All of the bands were reamplified under the same conditions in which they were generated. The fragments were cloned into the vector pGEM®-T Easy and the presence of the inserts was corroborated by colony PCR with the left and right universal oligonucleotides pUC/M13 and subsequently by restriction digest analysis, which revealed the expected size for each fragment. Only the 300-bp sequence showed 100% identity with C. immitis and C. posadasii with the sequences deposited in GenBank, whereas the sequences of the 250-, 180-, and 150-bp bands showed no identity with Coccidioides spp. sequences. The 300-bp sequence was subsequently named SCAR300.
Oligonucleotide design for the identification of Coccidioides spp.Using the SCAR300 marker, the specific oligonucleotides SCAR300 (F) (5′-AATGGCGTTAAGTGGGTC-3′) and SCAR300 (R) (5′-AAGCCACTTACACAATCCAG-3′) were designed. PCR was performed in a 25-µL reaction containing 10 ng of DNA, 2.0 mM MgCl2, 200 µM deoxynucleotide triphosphates (Applied Biosystems, Foster City, California, USA), 0.1 nmol of each oligonucleotide (SCAR300-F and SCAR300-R), and 1 U of Taq DNA polymerase (Applied Biosystems) in 1 × PCR buffer (Applied Biosystems). The amplification conditions were as follows: one cycle at 94 °C for 5 min; 30 cycles at 94 °C for 30 s, 53 °C for 30 s, 72 °C for 1 min; and a final extension at 72 °C for 5 min. Gel electrophoresis was performed in a 1% agarose gel with 0.5× Tris-borate-EDTA at 100 V.
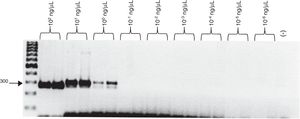
Evaluation of the sensitivity and specificity of the SCAR, Ag2/PRA, and microsatellite 621 and GAC2 markersThe minimum amount of DNA detected by PCR using the SCAR300 marker was 1 ng/µL (Fig. 1).
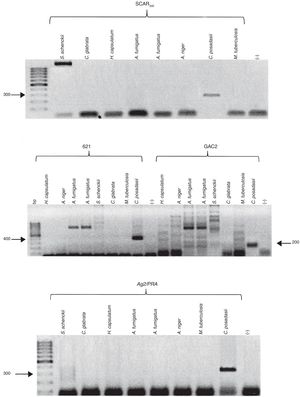
The SCAR300 marker was amplified using the C. posadasii (HU-1) DNA and exhibited the expected 300-bp product, whereas no amplification was observed using the DNA from the other assayed pathogenic fungi and M. tuberculosis (Fig. 2A). The expected 400-bp product was amplified for the 621 microsatellite using the C. posadasii (HU-1) DNA, whereas no amplification was observed using DNA from the other pathogenic fungi (Fig. 2B). The expected 200-bp fragment was amplified for the GAC2 microsatellite using the C. posadasii (HU-1) DNA, although additional products of different sizes were amplified when DNA from A. fumigatus, H. capsulatum, A. niger, S. schenckii, and M. tuberculosis was used (Fig. 2B). In addition, the expected 300-bp amplicon was obtained for the Ag2/PRA marker using C. posadasii (HU-1) DNA, although it a 200-300-bp product was also amplified using S. schenckii DNA (Fig. 2C).
Specificity of the molecular markers for the detection of Coccidioides spp. (A) SCAR300, (B) 621, GAC2 and (C) Ag2/PRA. The specificities of the molecular markers were determined as described in the Materials and Methods section using DNA from other pathogenic fungi and M. tuberculosis. Positive control (+); negative control (−); bp (molecular size marker).
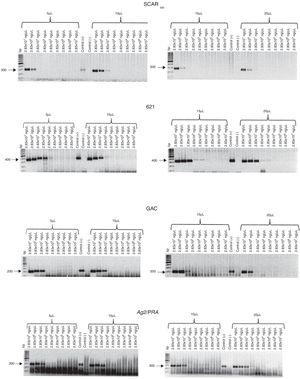
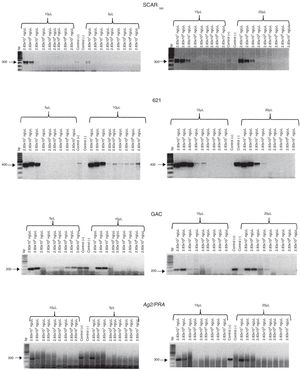
The concentrations of total DNA obtained from blood and sputum samples spiked with C. posadasii (HU-1) DNA were 10–100 and 50–100 ng/µL, respectively. The expected 300-bp band was observed when 10, 15, and 20 µL of total DNA was used from the blood spiked with 2.83 × 101 to 2.83 × 10−1 ng/µL of Coccidioides DNA, while the band was detected when 5 µL of total DNA was used from blood spiked with 2.83 × 101 ng/µL of Coccidioides DNA (Fig. 3). In contrast, for the sputum samples, the 300-bp band was observed when 5 and 15 μL of total DNA was used from sputum spiked with 2.83 × 101 to 2.83 × 100 ng/μL of Coccidioides DNA, while the amplicon was observed when 10 and 20 µL of total DNA was used from sputum spiked with 2.83 × 101 to 2.83 × 10−1 ng/µL of Coccidioides DNA (Fig. 4).
Sensitivity of the SCAR300, 621, GAC2, and Ag2/PRA markers. Different volumes (5, 10, 15, and 20 µL) of total DNA obtained from blood spiked with different concentrations of C. posadasii reference strain (HU-1) DNA were used. Positive control (+); negative control (−); bp (molecular size marker).
Sensitivity of the SCAR300, 621, GAC2, and Ag2/PRA markers. Different volumes (5, 10, 15, and 20 µL) of total DNA obtained from sputum spiked with different concentrations of C. posadasii reference strain (HU-1) DNA were used. Positive control (+); negative control (−); bp (molecular size marker).
When blood or sputum that was spiked with different concentrations of Coccidioides DNA harboring the GAC2 microsatellite was assayed for this marker by PCR, the expected 200-bp band was observed when five and 10 µL of total DNA was used from blood spiked with 2.83 × 101 to 2.83 × 10−6 and 2.83 × 101 to 2.83 × 10−1 ng/µL of Coccidioides DNA, respectively. In addition, the expected amplicon was observed when 15 and 20 µL of total DNA was used from blood spiked with 2.83 × 101 to 2.83 × 10−2 ng/µL of Coccidioides DNA (Fig. 3). For the sputum, a 200-bp amplicon was observed when 20 µL of total DNA was used from sputum spiked with 2.83 × 101 to 2.83 × 100 of Coccidioides DNA, while the expected band was observed when 5, 10, and 15 µL of total DNA was used from sputum spiked with 2.83 × 101 to 2.83 × 10−1 ng/µL of Coccidioides DNA (Fig. 4).
The 400-bp 621 microsatellite amplicon was observed when 5 µL of total DNA was used from blood spiked with 2.83 × 101 to 2.83 × 10−1 ng/µL of Coccidioides DNA, while the expected amplicon was observed when 10 µL of total DNA was used from blood spiked with 2.83 × 101 to 2.83 × 10−7 ng/µL of Coccidioides DNA. In contrast, the 400-bp amplicon was observed when 15 and 20 µL of total DNA was used from blood spiked with 2.83 × 101 to 2.83 × 10−3 and 2.83 × 101 to 2.83 × 10−2 ng/µL of Coccidioides DNA, respectively (Fig. 3). For the sputum samples, the 400-bp amplicon was observed when 5 and 10 µL of total DNA was used from sputum spiked with 2.83 × 101 to 2.83 × 10−7 and 2.83 × 101 to 2.83 × 10−6 ng/µL of Coccidioides DNA, respectively. In contrast the expected amplicon was observed when 15 and 20 µL of total DNA was used from sputum spiked with 2.83 × 101 to 2.83 × 10−3 ng/µL and 2.83 × 101 to 2.83 × 10−2 ng/µL of Coccidioides DNA, respectively (Fig. 4).
Evaluation of the Ag2/PRA marker with blood and sputum samplesFor the Ag2/PRA marker, the 300-bp band was observed when 5, 10, and 15 µL of total DNA was used from blood spiked with 2.83 × 101 to 2.83 × 100 ng/μL of Coccidioides DNA. In contrast, the expected amplicon was observed when 20 µL of total DNA was used from blood spiked with 2.83 × 101 to 2.83 × 10−1 ng/µL of Coccidioides DNA (Fig. 3). For the sputum samples, the 300-bp band was observed when 5, 10, 15, and 20 µL of total DNA was used from sputum spiked with 2.83 × 101 to 2.83 × 10−5, 2.83 × 101 to 2.83 × 10−3 ng/µL, 2.83 × 101 to 2.83 × 10−7 ng/μL, and 2.83 × 101 to 2.83 × 10−2 ng/µL of Coccidioides DNA, respectively (Fig. 4).
DiscussionVarious molecular markers have been described for the identification of Coccidioides spp. for diagnostic and epidemiological purposes.1,11,13,15,16,23 However, many of these markers have low sensitivity, specificity, and reproducibility as well as limitations associated with complicated methodologies that involve high costs. A small number of markers have been obtained from ribosomal genes, which are naturally conserved within the fungal kingdom; however, their use can lead to nonspecific results among several fungal species.24,25 In addition, commercially available probes used for diagnostic tests also yield nonspecific results in some cases.26 Due to these drawbacks, designing more specific and sensitive markers for the identification of Coccidioides was necessary. SCAR markers have been designed for other pathogenic fungi and are excellent candidates for this purpose, as described by Frías De León et al.20 Due to their high specificity and sensitivity, molecular methods are gradually being implemented as routine methods in clinical laboratories to confirm the information obtained through conventional methods. Furthermore, these newer techniques are used as auxiliary methods in the diagnosis of questionable cases of some mycoses as well as to facilitate the characterization of infection sources and to consolidate epidemiological information of such mycoses, especially in Latin American countries.
SCAR markers have proven to be very useful, with the development of a SCAR marker requiring the use of two specific primers that are designed from the nucleotide sequences of amplicons generated using techniques such as random amplification of polymorphic DNA (RAPD) or AFLP, after which they are cloned and associated with a feature of interest. Once developed, a SCAR marker can be applied to a large number of samples that can be simultaneously examined, reducing the necessary time and increasing reliability.27
It is important to mention that the development of molecular markers from native isolates, as is the case of the SCAR300 marker, is very important, since great genetic variability has been observed in isolates of C. immitis and C. posadasii from different geographic origins.24,28–30 Furthermore, it has been suggested that molecular markers used to detect pathogens from clinical samples should be designed from native isolates in the region in which they are to be used. Thus, it is important that the SCAR300 marker be validated for use in these countries.
Although one of the objectives of the present study was to design specific SCAR markers that were for C. immitis and C. posadasii, this was not possible due to the small number of isolates belonging to the C. immitis species and because of the high diversity found in all isolates, which made it difficult to identify species-specific bands in the polymorphic patterns obtained by AFLP. Thus, the designed marker was specific only at the genus level. However, because the SCAR300 marker was tested using isolates from Mexico and Argentina and showed good specificity and sensitivity, it may be a good candidate for the identification of fungi of the genus Coccidioides. However, it is necessary that the marker be evaluated with clinical samples and samples obtained from different sources of infection.
The results of this study showed that the SCAR300 marker was efficient at amplifying Coccidioides DNA from biological samples (blood or sputum) experimentally spiked with different concentrations of HU-1 strain DNA, even though it showed lower sensitivity than the GAC2 and 621 microsatellites.1 The SCAR300 marker was also shown to have a sensitivity that is highly similar to that of the marker Ag2/PRA, confirming its usefulness as a diagnostic tool for coccidioidomycosis (Fig. 3 and 4).
Regarding the specificity of the SCAR300 marker, it was shown to be as specific as the 621 microsatellite marker, since both were only amplified using DNA from C. posadasii. In contrast, in addition to DNA from C. posadasii, the GAC2 microsatellite was amplified using DNA from A. fumigatus, A. niger, H. capsulatum, S. schenckii, and M. tuberculosis, whereas the Ag2/PRA marker was amplified using DNA from C. posadasii and S. schenckii, making both of these markers less efficient for use in the diagnosis of coccidioidomycosis.
According to the results obtained in the present study with respect to the use of different volumes of total DNA (biological sample spiked with DNA from the HU-1 isolate), the use of 10 and 15 µL of total DNA from blood and sputum is recommended for the PCR with SCAR300 marker. This information is useful as a guide for laboratory personnel in charge of performing molecular diagnosis, since these results indicate the minimum volume of total sample DNA needed to perform PCR and achieve a positive result.
ConclusionsAlthough the 621 microsatellite marker showed greater sensitivity than the SCAR300 marker, it is important to note that the latter presents the advantage of having been obtained from isolates from Mexico and Argentina, which ensures the detection of the fungus in these countries.
However, it is essential that the SCAR300 marker, which was tested in this study using biological samples spiked with DNA from a C. posadasii isolate (HU-1), be tested with the largest possible number of clinical samples from patients with suspected coccidioidomycosis to definitively validate the method and corroborate its diagnostic utility.
Conflict of interestAll the authors of this study declare the absence of any potential conflicts of interest.
This work was financially by PAPIIT-DGAPA (IN215509-3).