Describe the presence of CTX-M-1 phylogenetic subgroup extended-spectrum β-lactamases (ESBL), associated with TEM and SHV genes, and the gene encoding cephalosporinase, CMY-2 in Escherichia coli and Klebsiella pneumoniae isolates from community-acquired urinary tract infections.
Methods102 E. coli and 21K. pneumoniae were collected from patients with culture-proven urinary tract infection (UTI), during February and March, 2011. Antimicrobial susceptibility test was performed by disk diffusion according to the standards of the Clinical Laboratory Standard Institute. Screening for cephalosporins-resistant E. coli and K. pneumoniae was performed by PCR assay for blaTEM, blaSHV, blaCTX-M-1,-2,-8,-9, blaPER-2 and blaCMY-2 genes. Statistical analysis was performed by chi-squared test and multivariate logistic regression analysis.
ResultsESBL production was detected in 12 (11.7%) E. coli and four (19%) K. pneumoniae isolates. TEM ESBLs were detected in seven E. coli and three K. pneumoniae isolates. SHV ESBLs were found in four K. pneumoniae isolates. CTX-M-1 phylogenetic subgroup was positive in seven E. coli and three K. pneumoniae isolates. CMY-2 β-lactamase gene was detected in nine E. coli and one K. pneumoniae isolates. A significant association of ESBL expression in E. coli was observed with resistance to tobramycin (p≤0.001), tetracycline (p=0.043), and ciprofloxacin (p≤0.001). In K. pneumoniae isolates, significant association was found with resistance to tobramycin and ciprofloxacin (p=0.006), and trimethoprim-sulfamethoxazole (p=0.043). Multivariate analyses did not show association between ESBL production in E. coli and K. pneumoniae, and resistance to non-β-lactams drugs.
ConclusionsCTX-M ESBL in uropathogens isolated from the community is cause for concern due to the enormous potential for multidrug resistance from strains that produce these enzymes, which could lead to failure of empirically-administered therapies and development of complicated UTIs.
Infections caused by extended-spectrum β-lactamases (ESBLs)-producing E. coli and K. pneumoniae have been described in the hospital setting,1 although during the last decade these organisms have begun to disseminate into the community, becoming an emerging public health problem.2 ESBLs are enzymes commonly associated with TEM and SHV. However, many E. coli and K. pneumoniae isolates possess the gene encoding ampC cephalosporinase.3 Moreover, CTX-M-type ESBLs have emerged within the community, particularly among E. coli and K. pneumoniae isolated from urinary tract infections (UTIs), with a widespread prevalence and multidrug resistance in many countries worldwide.2,4 Multidrug resistance expressed by CTX-M producing isolates from the community is often associated with the presence of multiple ESBLs genes, as well as aminoglycoside and quinolone resistance genes,5,6 thereby limiting the choice of effective antimicrobial drugs.7
The aim of this study was to describe the presence of the CTX-M-1 phylogenetic subgroup ESBL associated with TEM and SHV genes and the gene encoding cephalosporinase, CMY-2, in E. coli and K. pneumoniae isolates from community-acquired UTIs.
Materials and methodsBacterial isolatesDuring a period of two months (February and March) in 2011, 123 isolates of enterobacteria (102 Escherichia coli and 21 Klebsiella pneumoniae), were collected from patients with culture-proven UTI evaluated in four clinical microbiology laboratories in Valledupar, Colombia. Cultures were obtained from midstream-collected urine. A uroculture was considered positive when it presented a bacterial count of more than 100,000 colony-forming units (CFU)/mL in a urine specimen collected midstream.8E. coli and K. pneumoniae identification was performed according to conventional method.9 This study was based on laboratory records and not on medicals records. Organisms were isolated from non-hospitalized patients during an outpatient visit to any of the four clinical microbiology laboratories.
This study did not require an approval from the Ethics Committee of Institute for Tropical Biological Research from Universidad de Córdoba, Colombia.
Susceptibility testing and ESBL detectionAntimicrobial susceptibility and interpretation were performed by disk diffusion according to Clinical Laboratory Standard Institute (CLSI).10 The antimicrobials used were: piperacillin-tazobactam (100μg/10g), ceftazidime (30μg), ceftriaxone (30μg), cefepime (30μg), aztreonam (30μg), cefoxitin (30μg), ertapenem (10μg), imipenem (10μg), tobramycin (10μg), ciprofloxacin (5μg), trimethoprim-sulfamethoxazole (25μg), and tetracycline (30μg) from Oxoid, Basingstoke, United Kingdom. ESBL phenotype was detected by interpretation of the CLSI criteria and disk combination method.11E. coli ATCC® 25922 and K. pneumoniae ATCC® 700603 were used as control strains.
PCR amplificationScreening of cephalosporins-resistant E. coli and K. pneumoniae isolates was performed by PCR assay for blaTEM, blaSHV, blaCTX-M-1, blaCTX-M-2, blaCTX-M-8, blaCTX-M-9, blaPER-2 and blaCMY-2 genes using the primers described elsewhere.12–16
Statistical analysisA statistical comparison of the frequencies of ESBL presence in E. coli and K. pneumoniae isolates was conducted by chi-squared test and multivariate logistic regression analysis. A value of p<0.05 was considered significant. All statistical calculations were performed by using the Statistical Analysis System version 9.1.3 (SAS, Cary, North Carolina, USA).
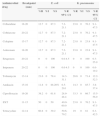
ResultsAntibiotic susceptibilitySusceptibility profile expressed by 123 enterobacteria isolates (102 E. coli and 21K. pneumoniae) is shown in Table 1. E. coli isolates included in this study presented resistance to broad-spectrum cephalosporins in 23.8% of cases. In K. pneumoniae the resistance rate to broad-spectrum cephalosporins was 12.7%. Resistance expressed by E. coli and K. pneumoniae isolates was of substantial level to non-β-lactam agents (Table 1). None of the strains were resistant to carbapenems.
Percentage of antibiotic susceptibility expressed by E. coli and K. pneumoniae isolates.
| Antimicrobial drug | Breakpoint (mm) | E. coli | K. pneumoniae | ||||||
|---|---|---|---|---|---|---|---|---|---|
| %R | %I | %S | %R 95% CI | %R | %I | %S | %R 95% CI | ||
| Ceftazidime | 18-20 | 12.7 | 0 | 87.3 | 7.2-21.1 | 23.8 | 0 | 76.2 | 9.1-47.5 |
| Ceftriaxone | 20-22 | 12.7 | 0 | 87.3 | 7.2-21.1 | 23.8 | 0 | 76.2 | 9.1-47.5 |
| Cefepime | 15-17 | 12.7 | 0 | 87.3 | 7.2-21.1 | 23.8 | 0 | 23.8 | 9.1-47.5 |
| Aztreonam | 18-20 | 12.7 | 0 | 87.3 | 7.2-21.1 | 23.8 | 0 | 23.8 | 9.1-47.5 |
| Ertapenem | 20-22 | 0 | 0 | 100 | 0.0-4.5 | 0 | 0 | 100 | 0.3-25.9 |
| Imipenem | 20-22 | 0 | 0 | 100 | 0.0-4.5 | 0 | 0 | 100 | 0.0-19.2 |
| Tobramycin | 13-14 | 21.6 | 0 | 78.4 | 14.3-31.1 | 28.6 | 0 | 71.4 | 12.2-52.3 |
| Amikacin | 15-16 | 11.8 | 0 | 88.20% | 6.5-20.1 | 14.3 | 0 | 85.7 | 3.8-37.4 |
| Ciprofloxacin | 16-20 | 38.2 | 0 | 61.8 | 28.9-48.4 | 33.3 | 0 | 66.7 | 15.5-56.9 |
| SXT | 11-15 | 50 | 0 | 50 | 40.0-60.0 | 23.8 | 0 | 76.2 | 9.1-47.5 |
| Tetracycline | 12-14 | 60.8 | 0 | 39.2 | 50.6-70.2 | 19 | 0 | 81 | 6.3-42.5 |
R, resistance; I, intermediate resistance; S, susceptible; SXT, trimethoprim-sulfamethoxazole; CI, confidence interval.
Eighteen broad-spectrum cephalosporins-resistant isolates were tested by disk combination method, and ESBL production was found in 11 E. coli (10.7%) and four K. pneumoniae (19%) isolates by phenotypic confirmation test, as the zone of inhibition increased in presence of clavulanic acid. Three isolates showed full resistance to third-generation cephalosporins and to a combination of both cefotaxime and ceftazidime with clavulanic acid, a phenotype consistent with AmpC β-lactamase production (Table 2).
Molecular and phenotypic characteristics of third-generation cephalosporins-resistant E. coli and K. pneumoniae isolates.
| Pathogen | No. isolates | Disk combination method | ESBL genes | Susceptibility by disk diffusion CLSI | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| blaTEM | blaSHV | blaCTX-M-1 | blaCMY-2 | AK | FOX | CIP | ETP | IPM | TZP | TCY | TOB | SXT | |||
| E. coli | |||||||||||||||
| 35 | + | + | - | + | + | S | S | R | S | S | S | R | R | S | |
| 40 | + | - | - | + | + | S | S | R | S | S | R | R | R | R | |
| 102 | + | + | - | - | + | S | R | R | S | S | S | S | S | S | |
| 108 | + | - | - | - | - | S | S | R | S | S | R | R | S | R | |
| 525 | - | - | - | - | + | R | R | R | S | S | R | R | R | R | |
| 527 | + | + | - | + | + | S | S | R | S | S | S | R | S | R | |
| 551 | + | + | - | + | + | S | S | R | S | S | S | R | R | R | |
| 576 | + | - | - | - | - | S | S | R | S | S | S | R | S | S | |
| 580 | - | - | - | + | + | S | R | R | S | S | R | R | R | R | |
| 612 | + | - | - | + | + | S | S | R | S | S | S | R | R | R | |
| 673 | + | + | - | - | - | S | S | R | S | S | R | R | R | R | |
| 678 | + | + | - | + | + | R | R | R | S | S | R | R | R | R | |
| 698 | + | + | - | - | + | S | S | R | S | S | S | R | R | R | |
| K. pneumoniae | |||||||||||||||
| 111 | + | - | + | + | - | S | S | R | S | S | R | S | R | R | |
| 587 | + | + | + | + | - | S | S | R | S | S | S | R | R | R | |
| 672 | - | - | - | - | + | R | R | S | S | S | R | S | S | S | |
| 710 | + | + | + | + | - | R | S | R | S | S | R | S | R | R | |
| 5125 | + | + | + | - | + | R | R | R | S | S | R | S | R | S | |
AK, amikacin; FOX, cefoxitin; CIP, ciprofloxacin; ETP, ertapenem; IPM, imipenem; TZP, piperacillin/ tazobactam; TCY, tetracycline; TOB, tobramycin; SXT, trimethoprim-sulfamethoxazole; R, resistance; S, susceptible; +, positive; -, negative.
Molecular analysis by amplification of purified DNA in 18 cephalosporins-resistant isolates yielded a PCR product with specific primers for blaTEM, blaSHV and blaCTX-M-1 genes in 16 isolates. The molecular method found one additional ESBL-positive E. coli isolate that the phenotypic method had not detected (Table 2). Therefore, ESBL production was detected in 12 (11.7%) E. coli isolates; blaCTX-M-2, blaCTX-M-8, blaCTX-M-9, and blaPER-2 were not detected by PCR screening. TEM ESBLs were detected in ten ESBL producers (seven E. coli and three K. pneumoniae). SHV ESBLs were found in four K. pneumoniae isolates. PCR amplification by CTX-M type ESBL was positive for CTX-M-1 phylogenetic subgroup in seven E. coli and three K. pneumoniae. CMY-2 β-lactamase gene was detected in nine E. coli and one K. pneumoniae isolates. A combination of ESBLs and AmpC β-lactamase genes was found in ten isolates (Table 2).
Four E. coli isolates were carrying both blaTEM and blaCTX-M-1 ESBLs genes, and all were positive for blaCMY-2. Two of four ESBL producing K. pneumoniae was carrying blaTEM, blaSHV and blaCTX-M-1 genes, another isolate was a producer of ESBL types SHV and CTX-M-1 subgroup, and one of the four isolates (5125) carrying blaTEM also contained the blaSHV gene, this being a single K. pneumoniae isolate positive for blaCMY-2 (Table 2).
A significant association of ESBL expression in E. coli was observed with resistance to tobramycin (p≤0.001), tetracycline (p=0.043), and ciprofloxacin (p≤0.001) (Table 3). A positive test result in K. pneumoniae isolates was significantly associated with resistance to tobramycin and ciprofloxacin (p=0.006), and trimethoprim-sulfamethoxazole (p=0.043) (Table 3). Multivariate analyses did not show any association between ESBL production in E. coli and K. pneumoniae, and resistance to non-β-lactams drugs (Table 4).
Crude association between antimicrobial drug resistance and positive test result for ESBL in E. coli and K. pneumoniae.
| Antimicrobial agent | E. coli (n=102), OR (95% CI) | p | K. pneumoniae (n=21), OR (95% CI) | p |
|---|---|---|---|---|
| Amikacin | 0.65 (0.07-5.56) | 0.932 | 16 (0.95-267) | 0.140 |
| Tobramycin | 10.85 (2.87-40.99) | < 0.001 | 55.8 (2.24-1,386.31) | 0.006 |
| Tetracycline | 8.41(1.04-67.94) | 0.043 | 1.55 (0.11-20.61) | 0.710 |
| Ciprofloxacin | 57.7 (3.29-1,009.91) | < 0.001 | 55.8 (2.24-1,386.31) | 0.006 |
| SXT | 3.42 (0.87-13.50) | 0.124 | 22.5 (1.50-335.33) | 0.043 |
OR, odds ratio; CI, confidence interval; SXT, trimethoprim-sulfamethoxazole.
Variables with p<0.05 were considered significant.
Association of antimicrobial drug resistance and presence of ESBL in E. coli and K. pneumoniae.
| Antimicrobial drug | Estimate | OR (95% CI) | p* |
|---|---|---|---|
| E. coli | |||
| Tetracycline | -0.23 | 0.0002 (0.0000-1.2730) | 0.812 |
| Ciprofloxacin | -0.30 | 0.0001 (0.0000-5.3914) | 0.762 |
| Tobramycin | -1.55 | 0.283 (0.058-1.383) | 0.119 |
| SXT | 0.19 | 1.230 (0.159-9.509) | 0.842 |
| K. pneumoniae | |||
| Tetracycline | 0.54 | 7.877 (0.004-12678.32) | 0.583 |
| Ciprofloxacin | -1.64 | 0.009 (0.00004-2.4168) | 0.100 |
| Tobramycin | -1.24 | 0.021 (0.00005-0.0407) | 0.212 |
| SXT | -0.54 | 0.119 (0.00005-253.05) | 0.586 |
SXT, trimethoprim-sulfamethoxazole; OR, odds-ratio; CI, confidence interval.
Community-acquired UTIs caused by ESBL-producing organisms are a cause of concern due to the clinical failure of empirical treatment protocols. This study shows two main pathogens causing UTIs in the community (E. coli and K. pneumoniae). The results show that expressed resistance to quinolones (ciprofloxacin) is similar among community isolates of E. coli and K. pneumoniae, 33% and 38%, respectively (Table 1). However, significant resistance was presented to trimethoprim-sulfamethoxazole (50%) and tetracycline (60%) in E. coli isolates, characterizing an alarming resistance to these two families of antibiotics.
ESBL production found among community isolates of E. coli was 11.7% and 19% for K. pneumoniae, rates similar to that found in hospital isolates;1,17K. pneumoniae is the main ESBL-producing organism worldwide.18 In Latin American countries, community-acquired ESBLs are starting to appear with rates up to 28.7% among E. coli and K. pneumoniae isolates,19 while in Europe reported rates are of only 1.5% to 5.6%, respectively.20 The presence of ESBL-producing urinary isolates in the community is a significant public health problem that unavoidably increases health care costs for patients and health care providers due to the use of carbapenems, since the oral agents (nitrofurantoin and fosfomycin), used for treatment of UTIs, are unsuitable due to the enormous resistance-development ability demonstrated by uropathogens, which has led to a loss of interest in these drugs. Therefore, carbapenems appear to be the drugs of choice for the treatment of UTIs caused by ESBL-producing E. coli and K. pneumoniae, considering their rational use.21,22
In Colombia, the prevalence of community-acquired ESBL-producing E. coli and K. pneumoniae is not well known; the findings of TEM and SHV enzymes in this study show the spread of these enzymes in hospital and community isolates in Colombia.23 However, the presence of community-acquired CTX-M-producing Enterobacteriaceae (seven E. coli and three K. pneumoniae), is of concern especially in E. coli, because these enzymes are commonly encoded by plasmids. Besides, there is the possibility that isolates found in this study are associated with well-characterized ST131 and ST405, which are multidrug-resistant intercontinental clones responsible for the dissemination of blaCTX-M-15, as previously reported in Colombia.24 Multidrug-resistance expressed by ESBL-producing Enterobacteriaceae in this study was presented to aminoglycosides, trimethoprim-sulfamethoxazole, tetracycline, and quinolones (Table 2).
ESBL-producing K. pneumoniae strains are more related to hospital outbreaks. However, the current distribution of ESBL carrying microorganisms in patients with community-acquired UTIs is a generalized problem, thus producing a flow of isolates from the community to the hospital setting.25 Likewise, the strains of E. coli and K. pneumoniae that harbored multiple ESBL genes are important, because it has been reported that isolates carrying multiple ESBL genes had a greater resistance to various classes of antibiotic mediated through acquisition of genes located on mobile genetic elements such as plasmids, transposons, or integrons.5,26 Furthermore, the fact that they harbor multiple ESBL genes in combination with the gene encoding cephalosporinase CMY-2 could explain the cefoxitin resistance (four E. coli and two K. pneumoniae), as reported in previous studies that have shown the blaCMY-2 gene linked downstream with the promoter sequence ISECP1,27 increasing the likelihood of therapeutic failures and difficulty for laboratory phenotypic detection of ESBL.28,29Finally, resistance expressed by E. coli and K. pneumoniae isolates, and molecular confirmation of ESBL genes were analyzed using statistical models to measure the crude association between a positive result for ESBL in isolates of E. coli and resistance to tobramycin (p≤0.001), tetracycline (p=0.025) and ciprofloxacin (p=0.01) (Table 3). In K. pneumoniae isolates, an association was found between a positive result for ESBL and resistance to tobramycin (p=0.002), ciprofloxacin (p=0.002), and trimethoprim-sulfamethoxazole (p=0.027) (Table 3). However, in multivariate analysis did not show non-β-lactam drugs as an independent variable. This finding suggests that non-β-lactams resistance remained independently associated with ESBL, but for reasons unrelated to ESBL genes. Studies have shown that non-β-lactam and β-lactam (ESBL) resistance encoding genes are located in plasmids.13,30 This common location may explain the resistance to previously mentioned antibiotics. Nevertheless, it is interesting that resistance to ciprofloxacin was associated with presence of ESBL and resistance to other antibiotic classes (Table 2). To some extent, this could be explained by its extensive use as an empirical drug for treatment of UTIs, resulting in strong selective antimicrobial pressure in the community. Similar data were found in previous studies.31–33
ConclusionDetection of CTX-M ESBL in uropathogens isolated from the community is of concern because of the enormous potential for multidrug resistance of strains that produce these enzymes, especially if E. coli isolates from this study could be related to the intercontinental clones ST131 and ST405, because empirically administered therapies will fail, which could lead to complicated UTIs.
Conflicts of interestAll authors declare to have no conflict of interest.
This work was supported by the Universidad de Córdoba, Institute for Tropical Biological Research.