There are several species of the genus Burkholderia, being the members of Burkholderia cepacia complex (Bcc) the main pathogens related to clinical infections in patients with cystic fibrosis (CF).1 Infection/colonization by Bcc in the airway is a cause of concern in CF as it may lead to a necrotizing pneumonia and clinical deterioration of individuals.1,2 Bcc species are often transmitted among CF patients therefore, the detection of these pathogens in the airways is relevant information to be considered for segregation of patients.1 The detection of microbial pathogens in the CF airways is traditionally performed by bacteriological culture3 but this method requires a quantity of viable bacterial cells in the clinical specimen. In contrast, culture independent methods, such as microbiome analysis which is based on Nucleic Acid Sequencing (16S rRNA), allows the identification of bacterial communities without the need of conventional culture.3
As a part of a more comprehensive study of microbiome analysis of sputum from CF patients; we conducted this study in order to compare the detection of the genus Burkholderia in the CF sputum by microbiome analysis and by the bacteriological culture.
A total of 22 sputa collected between July 2019 to March 2020 for routine bacteriological culture of 9 patients (5‒18 years old) from a CF reference hospital in southern Brazil, were submitted to microbiome sequencing. Patients had confirmed diagnosis of CF by the identification of the CFTR gene mutation. Bacteriological culture used selective media for Bcc and colonies with characteristics of Burkdolderia spp were submitted to Matrix-Assisted Laser Desorption/Ionization – Time of Flight Mass Spectrometry (MALDI-TOF MS) for final identification. For microbiome analysis, the DNA was extracted and was performed by 16S rRNA sequencing (V3V4 region) in an Illumina MiSeq (Illumina, San Diego, US). The bioinformatic analysis were performed with Bioconductor Workflow for Microbiome Data Analysis.4 DADA2 R package v1.16 algorithm and the SILVA version 138.1 were used for the determination of Amplicon Sequence Variants (ASV).
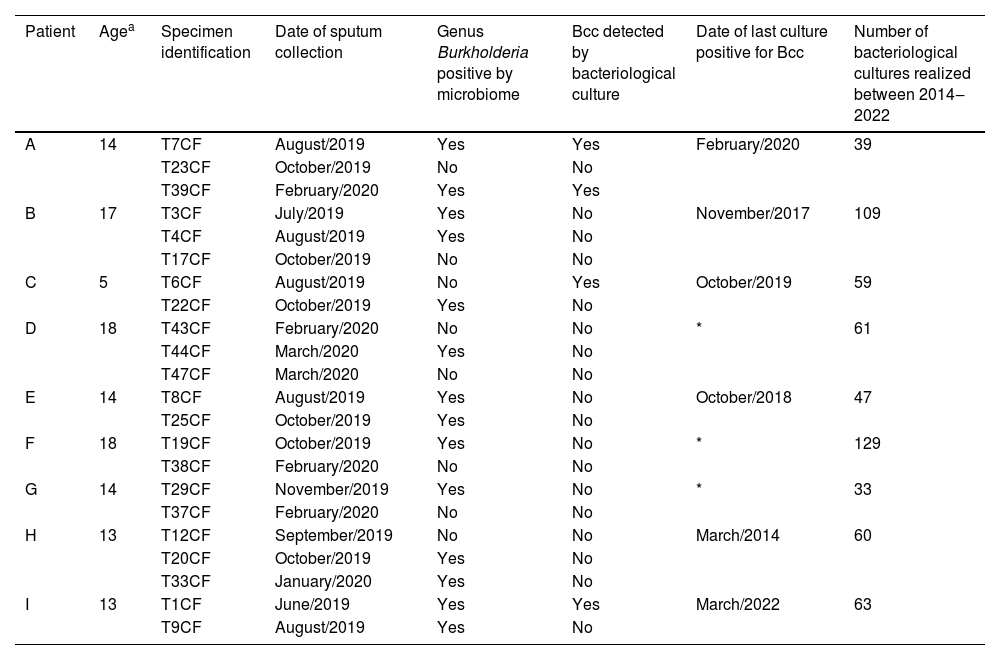
According to the microbiome analysis, it was possible to detect the genus Burkholderia in 14 specimens (63.6%) with at least one sputum specimen positive for all nine patients. Conversely, the bacteriological culture was capable to detect Bcc in only 4 sputa (18.2%) from 3 different patients. There was only one case (Patient “C”) of the same sputum (“T6CF” collected in August/2019) with bacteriological culture positive for Bcc and microbiome negative for Burkholderia spp. However, another sputum collected from Patient “C” two months later, presented the genus Burkholderia in the microbiome but no Bcc in the culture (Table 1). Noteworthy, three patients (D, F and G) never had Bcc identified by bacteriological culture, although, it was possible to detect the Burkholderia genus by microbiome. Analysis of previous data of bacteriological culture indicated that six of the nine CF patients included in this study presented Bcc in the sputum. The time lapse between the results of the culture positive for Bcc and the detection of Burkholderia spp by microbiome varied from 2 months to more than 5 years.
Comparison between the results of the bacteriological culture and microbiome analysis of 27 sputa from 9 CF patients considering the genus Burkholderia.
| Patient | Agea | Specimen identification | Date of sputum collection | Genus Burkholderia positive by microbiome | Bcc detected by bacteriological culture | Date of last culture positive for Bcc | Number of bacteriological cultures realized between 2014‒2022 |
|---|---|---|---|---|---|---|---|
| A | 14 | T7CF | August/2019 | Yes | Yes | February/2020 | 39 |
| T23CF | October/2019 | No | No | ||||
| T39CF | February/2020 | Yes | Yes | ||||
| B | 17 | T3CF | July/2019 | Yes | No | November/2017 | 109 |
| T4CF | August/2019 | Yes | No | ||||
| T17CF | October/2019 | No | No | ||||
| C | 5 | T6CF | August/2019 | No | Yes | October/2019 | 59 |
| T22CF | October/2019 | Yes | No | ||||
| D | 18 | T43CF | February/2020 | No | No | * | 61 |
| T44CF | March/2020 | Yes | No | ||||
| T47CF | March/2020 | No | No | ||||
| E | 14 | T8CF | August/2019 | Yes | No | October/2018 | 47 |
| T25CF | October/2019 | Yes | No | ||||
| F | 18 | T19CF | October/2019 | Yes | No | * | 129 |
| T38CF | February/2020 | No | No | ||||
| G | 14 | T29CF | November/2019 | Yes | No | * | 33 |
| T37CF | February/2020 | No | No | ||||
| H | 13 | T12CF | September/2019 | No | No | March/2014 | 60 |
| T20CF | October/2019 | Yes | No | ||||
| T33CF | January/2020 | Yes | No | ||||
| I | 13 | T1CF | June/2019 | Yes | Yes | March/2022 | 63 |
| T9CF | August/2019 | Yes | No |
Bcc, Burkholderia cepacia complex.
The data of this study indicated a high prevalence of the genus Burkholderia in sputum of CF patients according to microbiome when compared with the same specimens analyzed by bacteriological culture. Whether the identification of Burkholderia spp by microbiome analysis can predict the presence of Bcc in the airway of CF patients, it could be used to anticipate the treatment with antibiotics to prevent the increase of growth of the viable cells of the bacteria in the respiratory tract. It is important to avoid the growth of Burkholderia spp in the airway of CF for two main reasons: 1) To protect the individual patient from exacerbation of the respiratory disease associated with Bcc and 2) To decrease the transmission/circulation of Bcc among CF patients.
The eradication of Bcc was commonly accepted when the bacterium was not recovered by bacteriological culture over a follow-up period of at least 1 year with 3 sputum cultures negative.5 We found that, in three CF patients, Bcc was not identified by culture after more than one year, but their airway microbiome presented Burkholderia spp. This suggests that the concept of eradication of Bcc should be revised considering the information of 16S rRNA sequencing.
It is important to mention that the microbiome detected Burkholderia spp by 16S rRNA which may not guarantee the presence of viable bacteria in the specimen. Moreover, it is possible that the genus of Burkholderia as identified by microbiome does not include the species of Bcc related to the exacerbation of the respiratory disease in CF. However, one would be reckless to not consider the presence of the Burkholderia genus as an important information to be considered for the management of the CF respiratory disease. We consider that the data of both the bacteriological culture and microbiome analysis are complementary and should be considered in conjunction to contribute to the best clinical management of CF airway infections.
FundingThis work has been funded by INPRA ‒ Instituto Nacional de Pesquisa em Resistência Antimicrobiana ‒ Brazil (INCT/CNPq: 465718/2014-0 and INCT/FAPERGS: 17/2551-0000514-7) and by Fundo de Incentivo à Pesquisa e Eventos do Hospital de Clínicas de Porto Alegre (FIPE/HCPA) (Project n 2019-0659).