Studies have indicated that AMPK play critical roles in the regulation of innate immunity and inflammatory responses. However, the role of the polymorphisms of PRKAA1 gene in immune-response to infectious organisms remains unknown. To evaluate the potential role of PRKAA1/AMPKα1 in the immune-response to HBV, we conducted this case–control study.
MethodsWe recruited 276 patients (145 men and 131 women; average age, 51.6 years) with chronic HBV infection (CHB) and 303 healthy controls (166 men and 137 women; average age, 54.2 years). All the subjects were unrelated individuals of Chinese Han Population. Three SNPs of PRKAA1gene were tested.
ResultsRs1002424 polymorphism showed significant difference in the allele frequencies, but no difference in the genotype frequencies (allele: p=0.039411, OR95%CI=0.783479 [0.621067–0.988362]; genotype: p=0.104758); rs13361707 polymorphism showed significance in allele analysis, but not in genotype analysis (allele: p=0.034749, OR95%CI=1.284303 [1.017958–1.620335]; genotype: p=0.098027); rs3792822 polymorphism was demonstrated to have significant differences in both genotype and allele frequencies between cases and controls (allele: p=0.029286, OR95%CI= 0.741519 [0.566439–0.970716]; genotype: p=0.034560). The haplotype results showed that CTG and TCA in the rs13361707–rs1002424–rs3792822 block were significantly associated with the happening of HBV (CTG: p=0.036854, OR95%CI=1.281 [1.015–1.617]; p=0.030841, OR95%CI=0.743 [0.568–0.973]).
ConclusionThese findings suggest that PRKAA1 polymorphisms may contribute to the susceptibility of chronic HBV infection in Chinese Han origin.
Adenosine-monophosphate-activated protein kinase (AMPK) is highly conserved hetero-trimeric serine/threonine protein kinase consists of a catalytic subunit (α1 or α2) and two regulatory β (β1–2) and γ subunits (γ1–3).1,2 It is ubiquitously expressed and served as a master sensor of cellular energy homeostasis at the cellular and even whole-body level. AMPK is activated by the increase of AMP/ATP ratio from environmental or nutritional stress factors including oxidants, hypoxia and nutrient deprivation,3,4 then the activated kinase phosphorylates and regulates a variety of downstream molecules that maintains the balance between the consumption and generation of cellular energy.5,6 Disruption of this balance results a number of diseases such as diabetes, obesity, cardiovascular dysfunction, inflammation and some malignant diseases.7–9 In the last several years, accelerating evidence have indicated that AMPK plays critical roles in the regulation of innate immunity and inflammatory responses. Galic et al. reported that target deletion of AMPK β1 in mice enhanced adipose tissue macrophage inflammation and liver insulin resistance.10 While some other researchers demonstrated that AMPK-α1 is essential in mediating TLR4 or TNF-α triggered inflammatory signals as an activating kinase of TAK1.11,12 AMPK-α1 is encoded by PRKAA1 gene which is located on chromosome 5p12.1 and constitutes a 39kb region. Studies have indicated that nucleotide polymorphisms in PRKAA1 gene was significant associated with the risk of gastric, colon and rectal cancer.13–16 However, the role of PRKAA1 gene in the immune-response to infectious organisms remains unknown.
Hepatitis B is one of the most common infectious diseases caused by the hepatitis B virus (HBV) which attacks the liver. Chronic hepatitis B virus infection (CHB) may lead to liver fibrosis and cirrhosis, and dramatically increase the incidence of hepatocellular carcinoma (HCC). The susceptibility and progression of this disease are known to be influenced by viral load, virus genotype, environmental factors, host immune responses and host genetic factors.17,18 Considering the importance of PRKAA1 in the inflammatory signaling cascade, we conducted this case–control study to investigate the association between PRKAA1 and the susceptibility of chronic HBV infection in Chinese Han population.
Materials and methodsStudy subjectsIn this study, we recruited 276 patients (145 men and 131 women; average age, 51.6 years) with chronic HBV infection and 303 healthy controls (166 men and 137 women; average age, 54.2 years) from Shaanxi Province People's Hospital between Jan 2012 and Dec 2014. All the subjects were unrelated individuals of Chinese Han Population. The patients were diagnosed based on serological positivity for HBsAg, HBeAg or anti-HBe and anti-HBc for at least 6 months, and persistent abnormality of ALT (alanine aminotransferase) and AST (aspartate aminotransferase). All the patients were in the active hepatitis stage without evidence of liver cirrhosis and HCC. Patients with other diseases were excluded from this study. Healthy controls were randomly selected from healthy persons under routine health screening at the same hospital during the same time period. They were confirmed to be negative for HBsAg and HBeAg, without clinical evidence of hepatic disease, cancer and other disease. The main characteristics of the subjects are listed in the supplementary Table S1. Written informed consent was obtained from each participant of study. The study protocol conformed to the 1975 Declaration of Helsinki and was approved by the Research Ethics Committee of Shaanxi Province People's Hospital.
Genotyping of PRKAA1 gene polymorphismsGenomic DNA was isolated from EDTA peripheral blood using the DNA Extraction Kit (Tiangen Biotech Co., Ltd., Beijing, China) following the manufacturer's instructions. All DNA samples were amplified for rs13361707, rs1002424 and rs3792822 by PCR (polymerase chain reaction) using TaKaRa PCR Amplification Kit (Takara Bio Inc.). The primers were designed using primer 5.0 software (Molecular Biology Insights) according to the sequence from NCBI. The primers for rs13361707 primers were: Forward: 5′-ACGTGTTAAGGAAATAGC-3′, and Reverse: 5′-AACTGTGTGTATAGTGCAGG-3′. The primers for rs1002424 were: Forward: 5′-AGTTTGGAAGTTATCAGATC-3′, and Reverse: 5′-ACAGGTGTGAGCCACAGCAC-3′. The primers for rs3792822 were: Forward: 5′-ACTAGCATCAAAATGTCAGC-3′, and Reverse: 5′-GACTAGCAAGGCCTAGATCT-3′. The PCR products were then sequenced and analyzed by ABI 3700 DNA automated sequencer (Applied Biosystems) according to the standard protocol.
Statistical analysisData management and statistical analyses were conducted by STATA software 11.0 version for Windows (StataCorp LP, College Station, TX, USA). Hardy–Weinberg equilibrium was tested using the χ2 test. Between-group variables were compared by unpaired Student's t-test or Mann–Whitney U test where appropriate, and genotype/allele frequencies were compared by χ2 test. The linkage disequilibrium and haplotype construction were performed from the observed genotypes using SHEsis method (http://www.nhgg.org/analysis), and odds ratios (OR) with 95% confidence interval (CI) were calculated. A two-tailed p<0.05 was accepted as statistically significant.
ResultsSingle-point association analysisThere were no deviations from Hardy–Weinberg equilibrium for all studied polymorphisms in cases and controls (p>0.05). Totally, three SNPs of PRKAA1gene were tested. Rs1002424 polymorphism showed difference in the allele frequencies (p=0.039411, OR95%CI=0.783479 [0.621067–0.988362]) but no differences in the genotype frequencies (p=0.104758); Rs13361707 polymorphism showed significance in allele analysis (p=0.034749, OR95%CI=1.284303 [1.017958–1.620335]), but not in genotype analysis (p=0.098027); rs3792822 polymorphism was demonstrated to have significant differences in both genotype and allele frequencies between cases and controls (allele: p=0.029286, OR95%CI=0.741519 [0.566439–0.970716]; genotype: p=0.034560) (Table 1).
Distribution of genotype and allele frequencies of three SNPs in the PRKAA1 in HBV patients and healthy controls.
| SNP | HBV patients | Healthy controls | OR | 95%CI | p Value |
|---|---|---|---|---|---|
| (n=276) | (n=303) | ||||
| rs13361707 | |||||
| Allele(freq) | 1.284303 | 1.017958–1.620335 | 0.034749 | ||
| C | 269 (0.491) | 259 (0.429) | |||
| T | 279 (0.509) | 345 (0.571) | |||
| Genotype(freq) | 0.098027 | ||||
| CC | 71 (0.259) | 57 (0.189) | |||
| CT | 127 (0.464) | 145 (0.480) | |||
| TT | 76 (0.277) | 100 (0.331) | |||
| HWE | 0.228904 | 0.729947 | |||
| rs1002424 | |||||
| Allele(freq) | 0.783479 | 0.621067–0.988362 | 0.039411 | ||
| C | 278(0.507) | 343 (0.568) | |||
| T | 270(0.493) | 261 (0.432) | |||
| Genotype(freq) | 0.104758 | ||||
| CC | 76 (0.277) | 99 (0.328) | |||
| CT | 126 (0.460) | 145 (0.480) | |||
| TT | 72 (0.263) | 58 (0.192) | |||
| HWE | 0.184879 | 0.706059 | |||
| rs3792822 | |||||
| Allele(freq) | 0.741519 | 0.566439–0.970716 | 0.029286 | ||
| A | 121 (0.221) | 167 (0.276) | |||
| G | 427 (0.779) | 437 (0.724) | |||
| Genotype(freq) | 0.034560 | ||||
| AA | 14 (0.051) | 17 (0.056) | |||
| AG | 93 (0.339) | 133 (0.440) | |||
| GG | 167 (0.609) | 152 (0.503) | |||
| HWE | 0.821795 | 0.079962 |
Bold numbers represent P-values (P<0.05).
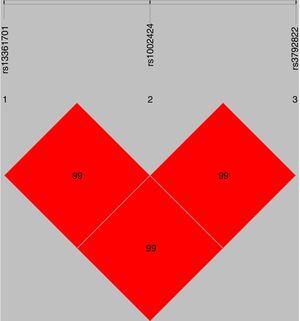
The standardized measure of linkage disequilibrium (LD), denoted as ‘D′’ was estimated at all possible pairs of SNP loci. LD for each pair of SNPs in cases and controls is presented in Table 2. The D′-value or r2-value showed strong linkage patterns were observed between rs13361707 and rs1002424 in all samples (D′=0.99), as well as among rs1002424 and rs3792822 (D′=0.99), and between rs13361707 and rs3792822 (D′=0.99) (Fig. 1).
Estimation of linkage disequilibrium between the 3 SNPs.
| rs13361707 | rs1002424 | rs3792822 | |
|---|---|---|---|
| rs13361707 | – | 0.996 | 1.000 |
| rs1002424 | 0.983 | – | 1.000 |
| rs3792822 | 0.282 | 0.285 | – |
SNP, single nucleotide polymorphism.
For each pair of SNPs, D′ values are shown above and r2 values below the diagonal, D’ or r2>0.9 are shown in boldface.
To facilitate identification of combinational effects of these three polymorphisms on the risk of chronic HBV infection, we employed haplotype analysis to study the frequency of the combination of multiple genetic variants. The results showed that CTG and TCA in the rs13361707–rs1002424–rs3792822 block were significantly associated with the development of chronic HBV infection (CTG: p=0.036854, OR95%CI=1.281 [1.015–1.617]; p=0.030841, OR95%CI=0.743 [0.568–0.973]) (Table 3).
Haplotype analysis: rs13361707, rs1002424, rs3792822.
| Case (freq) | Control (freq) | Pearson's p | χ2 | Odds ratio [95%CI] | |
|---|---|---|---|---|---|
| C T A | 0.01 (0.000) | 0.01 (0.000) | – | – | – |
| C T G* | 267.99 (0.489) | 258.99 (0.429) | 0.036854 | 4.358 | 1.281 [1.015–1.617] |
| T C A* | 120.99 (0.221) | 166.98 (0.276) | 0.030841 | 4.663 | 0.743 [0.568–0.973] |
| T C G* | 156.00 (0.285) | 176.02 (0.291) | 0.818643 | 0.053 | 0.971 [0.752–1.253] |
| T T A | 0.00 (0.000) | 0.01 (0.000) | – | – | – |
| T T G | 2.00 (0.004) | 1.99 (0.003) | – | – | – |
| C C G | 1.00 (0.002) | 0.00 (0.000) | – | – | – |
(All those frequency <0.03 will be ignored in analysis.)
Global χ2 is 5.885321 while df=2.
Pearson's p value is 0.052725.
Bold numbers represent P-values (P<0.05).
Chronic hepatitis B virus infection (CHB) is one of the most common infectious disease which leads to a huge economic problem on health care systems worldwide. According to the data from WHO, about 240 million people are chronically infected with hepatitis B and more than 780,000 people die from hepatitis B related diseases including cirrhosis and liver cancer each year (http://www.who.int/mediacentre/factsheets/fs204/en/). Thus it is important to clarify the mechanism of chronic HBV infection. Recent studies have shown that chronic hepatitis B virus infection is resulted from the interplay between the virus, host immune system and environmental factors.19,20 Among of them, host genetic backgrounds may be critical for the susceptibility and various outcomes of this disease.21,22 A number of genes have been identified to be associated with chronic hepatitis B virus infection by genome-wide association studies and case–control studies in the last several years.23–25 More knowledge on the understanding of candidate genetic factors may provide more clues to the prevention and therapy of this disease.
It has been well documented that AMPK is an important regulator of energy-sensing and signaling cascade in mammalian cells.26 As a heterotrimeric complex serine/threonine protein kinase, AMPK can regulate a variety of other physiological functions such as the regulation of cell proliferation and polarity, tumorigenesis, autophagy and innate immune responses.27 However, the signaling properties are diverse depending on the different combination of subunit isoforms of AMPK in various cells.28 Some studies indicated that AMPK may restrict HBV replication through promotion of autophagic degradation, but the underlying mechanism remains to be elucidated.29 The α1 catalytic subunit of AMPK encoded by PRKAA1 gene, is mainly expressed in lymphocytes and macrophages. AMPKα1 has been demonstrated to play a pivotal role in inducing pro-inflammatory signals through the activation of TAK1 and NF-κB,30 but its role in chronic HBV infection remains unclear. So, this current study might be the first attempt to assess the association between polymorphisms in PRAA1 gene and chronic HBV infection. The results showed that all the three polymorphisms we tested were associated with the susceptibility of chronic HBV infection (rs3792822 in both the allele- and genotype-analyses; rs13361707 and rs1002424 in allele-analyses). Based on the value of D′, the following haplotype-analyses were done and the results showed that, in the block of ‘rs13361707–rs1002424–rs3792822′, C T G and T C A were associated with chronic hepatitis B virus infection, suggesting that AMPKα1 may play some critical roles in the regulation of the initial defense against hepatitis B virus.
Taken together, the results of our study indicated that PRKAA1 gene is associated with the development of chronic HBV infection in Chinse Han Population. Although replications with larger sample size and functional studies to further clarify the role that PRKAA1/AMPKα1 are needed, the current study gave some clues which is useful in better understanding the intrinsic relationship between PRKAA1/AMPKα1 and the pathogenesis of chronic HBV infection.
Conflicts of interestThe authors declare no conflicts of interest.
This work was supported by grants from National High Technology Research and Development Program 863 (2014AA022304); Shaanxi Province Natural Science Foundation Research Project (2014JM2-8201).