In the current study we aimed to find out the impact of cytokine-inducible Src homology 2 domain protein (CISH) gene polymorphisms on the risk of pulmonary tuberculosis (PTB) in a sample of Iranian population.
Materials and methodsPolymorphisms of CISH rs2239751, rs414171, and rs6768300 were determined in 200 PTB patients and 200 healthy subjects using T-ARMS-PCR or PCR-RFLP method.
ResultsThe results showed that rs414171 A>T genotypes significantly decreased the risk of PTB (OR=0.16, 95% CI=0.10–0.27, p<0.0001, AT vs AA; OR=0.31, 95% CI=0.14–0.68, p<0.0001, TT vs AA; OR=0.19, 95% CI=0.12–0.29, p<0.0001, AT+TT vs AA; OR=0.29, 95%CI=0.20–0.42, p<0.0001, T vs A). For rs6768300, the findings indicated that this variant decreased the risk of PTB (OR=0.52, 95% CI=0.33–0.82, p=0.005, CG vs GG; OR=0.57, 95% CI=0.38–0.87, p=0.012, C vs G). No significant association was observed between CISH rs2239751 polymorphism and risk/protection of PTB.
ConclusionOur findings indicated that CISH rs414171 and rs6768300 variants might be associated with protection from PTB.
Tuberculosis (TB), an infectious disease, is the leading cause of global morbidity and mortality mainly in Asia and Africa.1,2 Roughly one-third of the world's population is infected with TB; however, approximately 5–15% of those infected individuals will develop active TB during their lifetime, typically within the first 2–5 years after the initial infection. It has been estimated that 9.6 million (5.4 million men, 3.2 million women and 1.0 million children) new cases occurred in 2014 according to the global tuberculosis report from World Health Organization (WHO).3 Though environmental and social factors also contribute to susceptibility to TB, there is considerable evidence that host genetic factors play a key role in susceptibility to the disease.4–6 In humans, the cytokine-inducible Src homology 2-containing protein is encoded by the CISH gene, which is located on the short arm of chromosome 3 (3p21.3).7 It contains four exons, of which exons 2–4 encode the CISH protein which is an important negative regulator for inflammatory cytokine signaling.7,8 It has been proposed that the interleukin-2 (IL-2)-mediated immune response is essential for host defense against infectious pathogens and CISH play a key role in controlling IL-2 signaling.9
Few studies have investigated the impact of CISH polymorphisms on TB and found an association between the variants and the risk of TB.10–12 To the best of our knowledge, there is no report regarding the possible association between CISH gene polymorphisms and the risk of PTB in the Iranian population. Thus, the present study aimed to examine the possible associations between rs2239751, rs414171 and rs6768300 polymorphisms of CISH gene and susceptibility to PTB in a sample of the Iranian population.
Materials and methodsStudy populationTwo hundred patients diagnosed with PTB and 200 healthy subjects were enrolled in this case-control study. The study design and enrollment process have been described previously.13,14 The cases were selected from PTB patients admitted to a university-affiliated hospital (Bou-Ali Hospital, Zahedan, referral center for TB). Briefly, diagnosis of PTB was based on clinical symptoms, radiological evidence, and bacteriological investigations such as sputum Acid Fast Bacilli (AFB) smear positivity, culture, and response to anti-tuberculosis chemotherapy. All control subjects were unrelated adults selected through the population without signs, symptoms or history of TB and from the same geographical origin, as the patients with PTB.
The local ethics committee of the Zahedan University of Medical Sciences approved the project, and informed consent was obtained from all individual participants included in the study. Genomic DNA was extracted from the whole blood by the salting out method as described previously.15
GenotypingIn this study we designed a tetra-primer amplification refractory mutation system polymerase chain reaction (T-ARMS PCR) for detection of CISH rs2239751 and rs414171polymorphism.
For rs2239751 we used two external primers (FO: GGGAAGACTACTTCTCCCTTGCTGTCT, RO: GCTGATGTGGTAGCTGGGTGTATGAATA) and two internal primers (FI (C allele)): GAACAAAGTTTTAGACTGCTGCGCTCTAC, RI (A allele): TTCTAGGTACATGTGTGTGCCCGTTT.
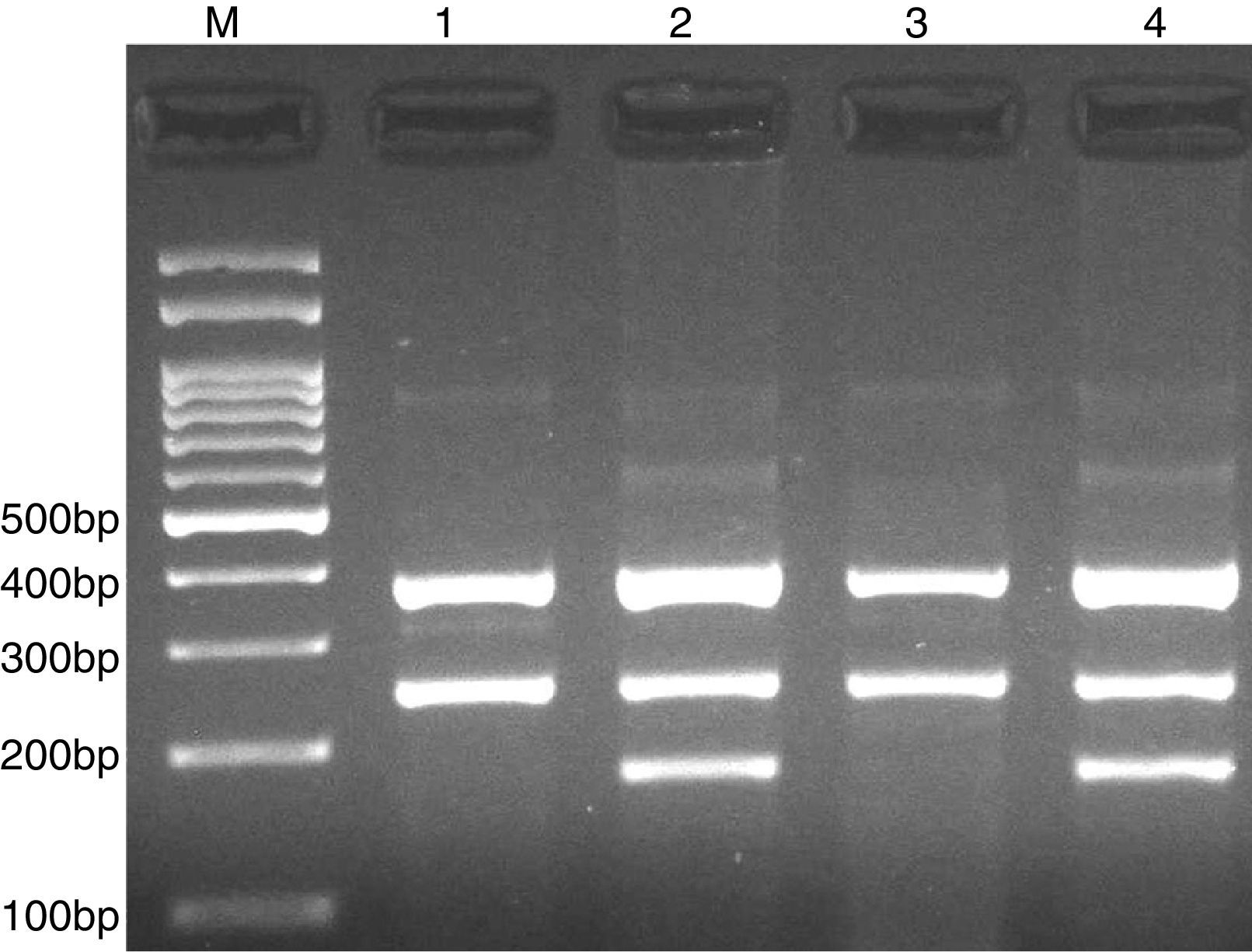
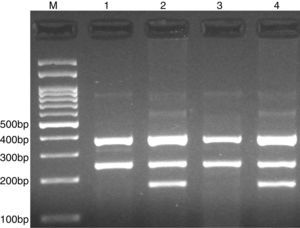
PCR was performed in 25μl reaction volumes containing 0.4μM of each primer, 250μM of each dNTP, 1U Taq DNA polymerase with 1.5mM MgCl2, and 50ng genomic DNA. The PCR cycling conditions was as follows: an initial denaturation step of 5min at 95°C followed by 30 cycles of 30s at 95°C, annealing at 62°C for 30s and extension at 72°C for 30s. Final extension was performed at 72°C for 5min. The PCR products were separated by electrophoresis in 2% agarose gels, and observed under ultraviolet light. Product sizes were C allele 180-bp, A allele 251-bp, and control 376-bp, as shown in Fig. 1.
For rs414171 we used two external primers FO: ACGCCGACAGACCTCCTTGGAGGAG, RO: GAAGCAGCGTCTTCCTAGAACCGCGG, and two internal primers FI (T allele): TGCTATTGGCCCTCCCCGACCACT, RI (A allele): CGCGACGCTGAAGGTGGAGCTGT.
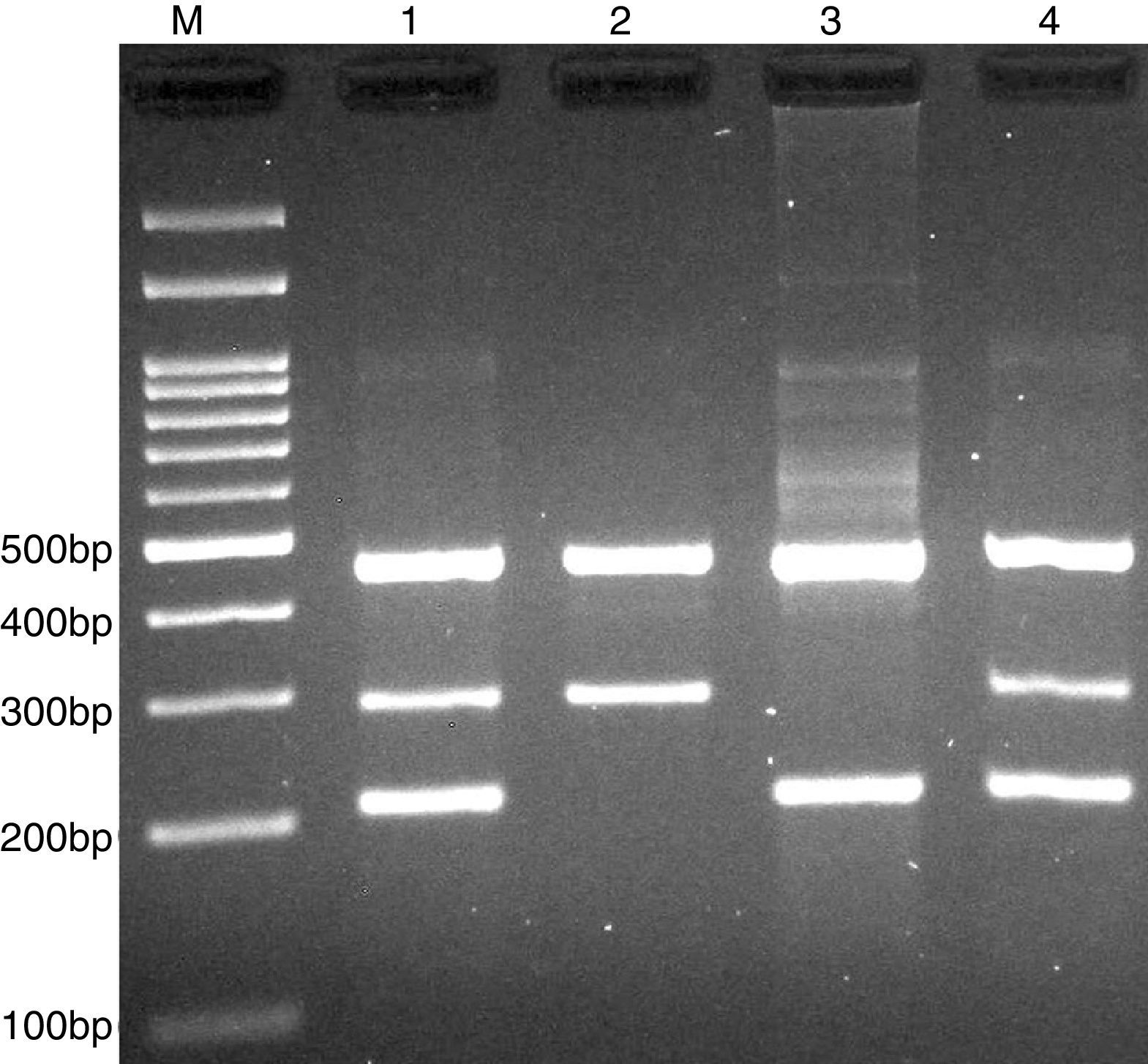
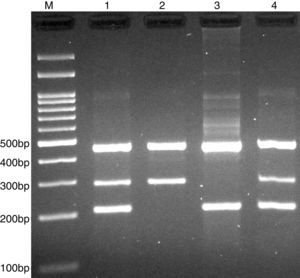
PCR was performed in 25μl reaction volumes containing 0.4μM of each primer, 250μM of each dNTP, 1U Taq DNA polymerase with 1.5mM MgCl2, and 50ng genomic DNA. The PCR cycling conditions was as follows: an initial denaturation step of 5min at 95°C followed by 30 cycles of 30s at 95°C, annealing at 68°C for 30s and extension at 72°C for 30s. Final extension was performed at 72°C for 5min. The PCR products were separated by electrophoresis in 2% agarose gels, and observed under ultraviolet light. Product sizes were A allele 290-bp, T allele 208-bp and control 452-bp, as shown in Fig. 2.
Detection of CISH rs6768300 polymorphism was done by PCR restriction fragment length polymorphism (PCR-RFLP) method. The set of forward and reverse primers were: 5′-GCGAGCTGCTGCCTAATC-3′ and 5′-GCTCGGCTCCACCTTCAG-3′, respectively.
PCR was performed in 25μl reaction volumes containing 0.4μM of each primer, 250μM of each dNTP, 1U Taq DNA polymerase with 1.5mM MgCl2, and 50ng genomic DNA. The PCR cycling conditions was as follows: an initial denaturation step of 5min at 95°C followed by 30 cycles of 30s at 95°C, annealing at 58°C for 30s and extension at 72°C for 30s. Final extension was performed at 72°C for 5min. PCR products were digested with HhaI restriction enzyme.
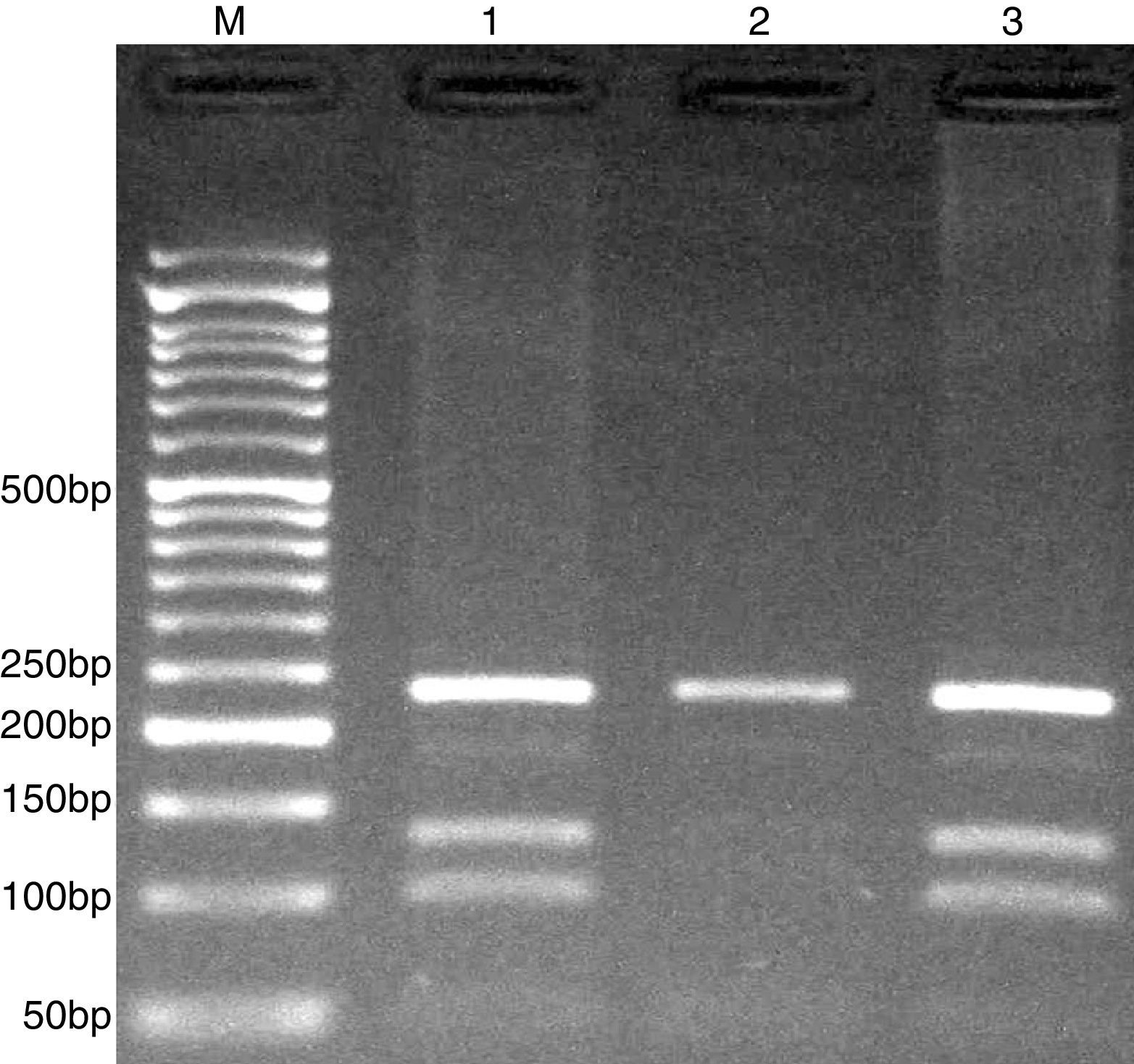
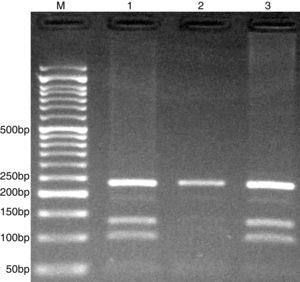
The PCR products were separated by electrophoresis in 2% agarose gels, and observed under ultraviolet light. G allele (undigested) 234-bp, while C allele digested and produces 101- and 133-bp (Fig. 3).
Statistical analysisAnalysis of the data was done using the SPSS 18.0 software. The differences between the variables were evaluated by chi-square test or independent sample t-test according to the data. The associations between genotypes and PTB were calculated by computing the odds ratio (OR) and 95% confidence intervals (95% CI) from logistic regression analyses. p-Value less than 0.05 was considered statistically significant.
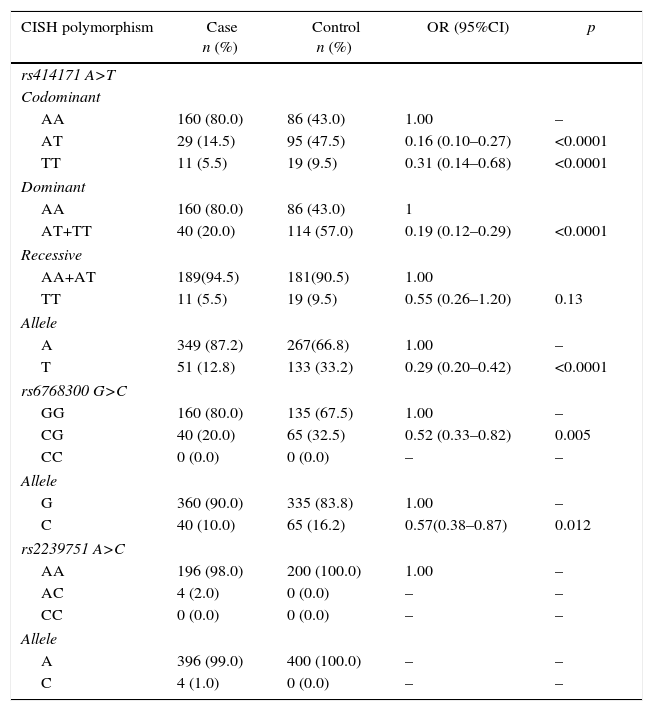
ResultsThe study consisted of 200 PTB (76 males, 124 females; ages 49.8±20.4 years) patients and 200 healthy subjects (87 males and 113 females; ages 49.2±15.1 years). There was no significant difference between the groups concerning sex and age (p=0.309 and p=0.707, respectively). Genotypes and allele frequencies of CISH polymorphisms are shown in Table 1.
Genotypes and allele frequencies of CISH polymorphisms in pulmonary tuberculosis (PTB) and control group.
| CISH polymorphism | Case n (%) | Control n (%) | OR (95%CI) | p |
|---|---|---|---|---|
| rs414171 A>T | ||||
| Codominant | ||||
| AA | 160 (80.0) | 86 (43.0) | 1.00 | – |
| AT | 29 (14.5) | 95 (47.5) | 0.16 (0.10–0.27) | <0.0001 |
| TT | 11 (5.5) | 19 (9.5) | 0.31 (0.14–0.68) | <0.0001 |
| Dominant | ||||
| AA | 160 (80.0) | 86 (43.0) | 1 | |
| AT+TT | 40 (20.0) | 114 (57.0) | 0.19 (0.12–0.29) | <0.0001 |
| Recessive | ||||
| AA+AT | 189(94.5) | 181(90.5) | 1.00 | |
| TT | 11 (5.5) | 19 (9.5) | 0.55 (0.26–1.20) | 0.13 |
| Allele | ||||
| A | 349 (87.2) | 267(66.8) | 1.00 | – |
| T | 51 (12.8) | 133 (33.2) | 0.29 (0.20–0.42) | <0.0001 |
| rs6768300 G>C | ||||
| GG | 160 (80.0) | 135 (67.5) | 1.00 | – |
| CG | 40 (20.0) | 65 (32.5) | 0.52 (0.33–0.82) | 0.005 |
| CC | 0 (0.0) | 0 (0.0) | – | – |
| Allele | ||||
| G | 360 (90.0) | 335 (83.8) | 1.00 | – |
| C | 40 (10.0) | 65 (16.2) | 0.57(0.38–0.87) | 0.012 |
| rs2239751 A>C | ||||
| AA | 196 (98.0) | 200 (100.0) | 1.00 | – |
| AC | 4 (2.0) | 0 (0.0) | – | – |
| CC | 0 (0.0) | 0 (0.0) | – | – |
| Allele | ||||
| A | 396 (99.0) | 400 (100.0) | – | – |
| C | 4 (1.0) | 0 (0.0) | – | – |
The results showed that rs414171 A>T genotypes significantly decreased the risk of PTB in codominant (OR=0.16, 95% CI=0.10–0.27, p<0.0001, AT vs AA; OR=0.31, 95% CI=0.14–0.68, p<0.0001, TT vs AA), and dominant (OR=0.19, 95% CI=0.12–0.29, p<0.0001, AT+TT vs AA) inheritance model tested. Furthermore, the T allele significantly decreased the risk of PTB (OR=0.26, 95% CI=0.18–0.38, p<0.0001) compared to A allele. For rs6768300, the findings showed that this CG genotype decreased the risk of PTB (OR=0.52, 95% CI=0.33–0.82, p=0.005) in comparison with GG genotype. The C allele significantly decreased the risk of PTB (OR=0.57, 95% CI=0.38–0.87, p=0.012) compared to G allele. No statistically significant association was detected between CISH rs2239751 polymorphism and risk/protective of PTB.
We estimated the Hardy–Weinberg equilibrium (HWE) for controls. In controls, the genotype distribution of rs414171 (χ2=0.982, p=0.321), but not rs6768300 variant (χ2=7.52, p=0.006) was in HWE. The rs2239751 variant was not polymorphic in our population.
DiscussionIn the present study we investigate the impact of CISH rs2239751, rs414171, and rs6768300 polymorphisms on the risk of PTB in a sample of the Iranian population. Our findings indicated an association between rs414171 and rs6768300 polymorphisms of CISH gene and risk of PTB. Regarding rs414171, the AT and TT genotypes as well as T allele significantly decreased the risk of PTB. For rs6768300, the CG genotype and G allele significantly decreased the risk of PTB. We found no significant association between rs2239751 variant and PTB.
In a case-control study, Ji et al.10 found that presence of either the T allele of rs414171 or the C allele of rs2239751 increased susceptibility to clinical TB in Chinese Han population. They found no significant association between CISH rs622502 variant and risk of TB.
Sun et al.12 investigated the impact of CISH gene promoter polymorphisms (rs414171, rs622502 and rs80945) on TB risk in Chinese children. They found that subjects carrying the rs414171TT homozygotes and rs809451GC heterozygotes had a 1.78-fold and 1.86-fold excess risk of developing TB compared to those with wild-type genotypes. Their findings proposed that CISH promoter rs414171 and rs809451 polymorphisms might play a vital role in mediating individual susceptibility to TB. Zhao et al.11 have found that rs2239751 and rs414171variants in CISH gene increased the risk of TB in Chinese Han population. They found no significant association between rs622502 and rs6768300 variants and TB risk.
Khor et al.9 investigated CISH polymorphisms at positions −639 (rs148685070), −292 (rs414171), −163 (rs6768300), +1320 (rs2239751), and +3415 (rs622502). They showed an association between CISH genetic variants and susceptibility to bacteremia, malaria, and tuberculosis when they considered all polymorphisms together in a multiple-SNP score.9
It has revealed that in humans the CISH gene is most consistently up-regulated by interleukin-2 stimulation,16 and it seems to be critical for T-cell proliferation and survival17 in response to infection. CISH controls the signaling pathway of a variety of cytokines, especially interleukin-2. CISH binds to the phosphorylated tyrosine residues of cytokine receptors and blocks the cytoplasmic docking and activation of signal transducer and activator of transcription 5 (STAT5),8,18–21 so inhibits downstream signal transduction.
One of the limitations of the present study is the relatively small sample size. There is no clear description for the deviation from HWE in our population. It may be due to the small sample size, genetic drift, or consanguineous marriages, which are common in this region of the country.
In summary, our finding emphasizes the impact of CISH polymorphisms on PTB protection in a sample of the Iranian population. Further studies with larger sample sizes and different ethnicities are warranted to confirm these findings.
Conflicts of interestThe authors declare no conflicts of interest.
This work was funded by a dissertation grant (MD thesis of AS) from Zahedan University of Medical Sciences. The authors thank the patients and healthy subjects who willingly participated in the study.