In response to the Zika epidemics in Brazil, the ZDC molecular assay (Bio-Manguinhos) was developed and registered at the Brazilian Regulatory Agency of Health Surveillance - ANVISA. The circulation of Zika (ZIKV) Dengue (DENV) and Chikungunya (CHIKV) viruses and their clinical similarities are challenges to correctly diagnose these viruses. The simultaneous detection of ZIKV, DENV and CHIKV is an important tool for diagnosis and surveillance. Here, we present the analytical and clinical performance evaluation of ZDC molecular assay (Bio-Manguinhos) at the public health laboratories three years after its registration at ANVISA. The clinical performance demonstrates the ZDC molecular assay (Bio-Manguinhos) has 100% sensitivity and 100% specificity to detect and discriminate ZIKV, CHIKV, and DENV from clinical plasma samples. The ZDC molecular assay (Bio-Manguinhos) results were highly reproducible and no cross-reactivity was seen during testing with a panel of other infectious agents. In conclusion, the ZDC molecular assay (Bio-Manguinhos) is an accurate and reliable tool to monitor Zika, dengue and chikungunya infections in countries like Brazil with simultaneous circulation of the three viruses.
The circulation of Zika (ZIKV) Dengue (DENV) and Chikungunya (CHIKV) viruses are responsible for significant epidemics around the world.1–3 Epidemiologically, ZIKV, CHIKV and DENV are mosquito-borne viruses (arboviruses) transmitted to humans by mosquitoes belonging to the Aedes genus. The Aedes mosquito is present worldwide posing a high risk for global transmission,4,5 with rapid geographic spread. ZIKV and DENV belongs to the family Flaviviridae, genus Flavivirus. The CHIKV belongs to the family Togaviridae, genus Alphavirus.3,6–9 While DENV and CHIKV are primarily transmitted through the bite of infectious mosquitoes, there are alternative means of human-to-human ZIKV transmission. ZIKV has been detected in urine and saliva, and there are case reports of sexual and perinatal transmission.1,3
ZIKV, DENV and CHIKV have similar clinical manifestations such as fever, myalgia and headaches at early stage of infection.2,10 The viral infection can be laboratory confirmed by viral RNA recognition or identification of serum specific antibodies.2,8,9,11 Virus culture and isolation is a very sensitive method, but not feasible to be used as a routine diagnostic tool because it is time-consuming and can require biosafety level 3 laboratory to reduce the risk of viral transmission. The cross-reactivity of antibodies between Flaviviruses, such as dengue, Zika, or yellow fever, limits the use of serology.12,13 Molecular tests are the most sensitive and discriminatory diagnostic tools for ZIKV, DENV or CHIKV,9,11 but the presence of nucleic acids in body fluids may be short-lived.14 It is recommended that real-time RT-PCR testing to be done within the first six days of the onset of illness.12,14
In 2016, in response to the ZIKV epidemics in Brazil, the ZDC molecular assay (Bio-Manguinhos) was developed and registered at the Brazilian Regulatory Agency of Health Surveillance - ANVISA. Clinical similarities and co-circulation of these arboviruses are challenges to correctly diagnose these viruses. The simultaneous detection of ZIKV, DENV and CHIKV is an important tool for diagnosis and vigilance through the public health laboratories. Transmission of the arboviruses ZIKV, CHIKV and DENV, in Brazil, can occur during the whole year, due to its tropical weather. Because of the concurrent arbovirus epidemics, and the overlapping endemic regions,10,15 the differential diagnosis must always include ZIKV, DENV and CHIKV infection.
Herein, we present the analytical and clinical performance evaluation of ZDC molecular assay (Bio-Manguinhos) three years after its registration at ANVISA and implementation at Brazilian public health laboratories.
Material and methodsZDC molecular assay (Bio-Manguinhos)The ZDC molecular assay (Bio-Manguinhos) is a real-time nucleic acid amplification, with an internal control (IC), designed to detect and discriminate in one reaction Zika/Chikungunya/IC and in the other reaction Dengue/IC, in samples previously submitted to nucleic acid extraction. The ZDC molecular can detect all serotypes of DENV, but do not differentiate among them. Primers and MGB-probes target NS1 gene of ZIKV (VIC), NSP1 gene of CHIKV (FAM), and 3’-NCR gene of DENV (VIC – DENV 2 and FAM – DENV 1, 3, 4), e IC (Dye3). The IC is a virus like particle protected by patent (PI0600715-5). The IC eliminates false-negative results and controls all steps of the reaction, from extraction to amplification, without interacting with the virus present in the plasma of infected patients. Positive controls supplied by the manufacturer are run in every batch. Additionally, an internal quality control (ICQ) from Bio-Manguinhos or a known positive sample of each laboratory of analysis is added to each run. The ICQ is a sample known to be positive for ZIKV, CHICK and DENV. The amplification is carried out on the Applied Biosystems 7500 Real-Time PCR System (Thermofisher) with the following conditions: 51 °C for 30 min, 95 °C for 10 min, and 40 cycles at 95 °C for 30 s and 58 °C for 30 s. A result was considered positive if any curve crossed this threshold prior to cycle 38 to Zika/Chikungunya and 33 to Dengue. Dengue results with Ct values between 33 and 37 are inconclusive, and the samples must be tested again.
Limit of detectionZIKV, CHIKV and DENV 1, 2, 3 and 4 RNA from virus culture were kindly provided by LAVIMOAN (Laboratório de Virologia Molecular, UFRJ) and LATEV (Laboratório de Tecnologia Viral Bio-Manguinhos) and were used to determine the LOD of the assay. Samples from the following reference panels were also used: (i) 1st World Health Organization International Standard for Zika virus RNA for Nucleic Acid Amplification Techniques (NAT)-Based Assays (PEI code 11468/16)16; (ii) 1st World Health Organization International Standard for Chikungunya virus RNA for Nucleic Acid Amplification Techniques (NAT)-Based Assays (PEI code 11785/16)17; (iii) Dengue Early Infection Accuset™ Performance Panel (Seracare 0845-0050). LOD was determined in eight independent assays at two-fold dilutions ranging 2.00E + 03 IU/mL to 2.50E + 01 ZIKV, 5.00E + 03 IU/mL to 8.00E + 00 to CHIKV and 3,90E + 04 to 1,22E+01to Dengue.
Additionally, LOD was also performed with a synthetic RNA that was synthetized by IDT (Integrated DNA Technologies, USA) to ZIKV and CHIKV containing the target sequence of the assay.
The software IBM SPSS Statistics, version 26, was used to perform the PROBIT analysis and determine LOD (95% confidence interval - 95% CI).
Specificity studiesThe specificity was evaluated by testing the detection of target and non-target viral RNA. Forty-four positive plasma samples previously tested by Trioplex real-time RT-PCR assay18 were used. Eleven samples were positive for ZIKV, seven for DENV and 26 for CHIKV. Ninety-six plasma samples previously tested by serological and/or molecular assays, and with negative results for Zika, Chikungunya and Dengue were used. Cross-reactivity with non-target was evaluated with samples previously tested and with positive results by serological and/or molecular assays to HIV, HCV, HBV, HTLV, Chagas disease, Yellow fever, Mayaro, and Syphilis.
Briefly, viral RNA was isolated from 500 µL of human plasma samples in the molecular biology workstation (BioRobot MDx, Qiagen) using the QIAamp one-for-all nucleic acid kit (Qiagen) in accordance with the manufacturer’s protocol. The internal control (IC) was added to each sample before extraction.
Clinical performanceIn order to evaluate the clinical performance of ZDC molecular assay (Bio-Manguinhos) 269 clinical samples were selected from the Ministry of Health's surveillance network. Fourty-one plasma samples were from Instituto Evandro Chagas (IEC), positive to ZIKV (n = 19), CHIKV (n = 13) and DENV (n = 9), previously tested by an in house protocol; and 228 plasma samples were from Feira de Santana, a city from Bahia state, previously collected from febrile cases in 2015 and without molecular diagnosis.
ReproducibilityA replicate panel was made spiking virus stock into truly negative plasma, to reach final Ct of 28 to ZIKV, CHIKV and DENV. Replicate tests were performed one week apart with different operators.
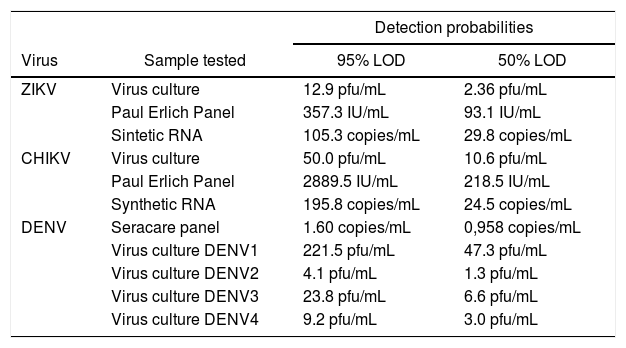
ResultsThe assessed results of analytical sensitivity with 95% and 50% positivity detection limits are summarized in Table 1.
Analytical sensitivity of ZDC molecular assay (Bio-Manguinhos).
| Detection probabilities | |||
|---|---|---|---|
| Virus | Sample tested | 95% LOD | 50% LOD |
| ZIKV | Virus culture | 12.9 pfu/mL | 2.36 pfu/mL |
| Paul Erlich Panel | 357.3 IU/mL | 93.1 IU/mL | |
| Sintetic RNA | 105.3 copies/mL | 29.8 copies/mL | |
| CHIKV | Virus culture | 50.0 pfu/mL | 10.6 pfu/mL |
| Paul Erlich Panel | 2889.5 IU/mL | 218.5 IU/mL | |
| Synthetic RNA | 195.8 copies/mL | 24.5 copies/mL | |
| DENV | Seracare panel | 1.60 copies/mL | 0,958 copies/mL |
| Virus culture DENV1 | 221.5 pfu/mL | 47.3 pfu/mL | |
| Virus culture DENV2 | 4.1 pfu/mL | 1.3 pfu/mL | |
| Virus culture DENV3 | 23.8 pfu/mL | 6.6 pfu/mL | |
| Virus culture DENV4 | 9.2 pfu/mL | 3.0 pfu/mL | |
The specific target detection was evaluated with plasma samples from febrile patients previously tested and confirmed ZIKV, DENV, CHIKV infections and negative (Table 2). No false positive was detected.
Performance of ZDC molecular assay (Bio-Manguinhos) with clinical plasma samples true positive and true negative.
| Specimen category | Tested | ZIKV positive | DENV positive | CHIKV positive | Sensitivity (95% CI) | Specificity (95% CI) |
|---|---|---|---|---|---|---|
| Zika | 11 | 11/11 | 0/11 | 0/11 | 100% (71.5– 100%) | 100% (97.2–100%) |
| Dengue | 7 | 0/7 | 7/7 | 0/7 | 100% (59–100%) | 100% (97.3–100%) |
| Chikungunya | 26 | 0/26 | 0/26 | 26/26 | 100% (86.8–100%) | 100% (96.2–100%) |
| Negative | 96 | 0/96 | 0/96 | 0/96 | N/A | N/A |
N/A not applicable.
Results of the clinical performance with ZDC molecular assay (Bio-Manguinhos) showed 100% positive agreement when testing IEC samples, with medium Ct of 33 to ZIKV, 22.7 to CHIKV and 22.5 to DENV. The evaluation of Feira de Santana samples have found 44 samples positive to CHIKV with mean Ct of 28.4, and six samples positive to ZIKV with mean Ct of 32.6.
The reproducibility within the laboratory was tested with a replicate panel with 100% agreement. All positive samples were detected and there was no false positive result. The mean Ct (+ SD) was 28.28 (+ 0.32) to ZIKV, 27.16 (+0.87) to CHIKV, 25.47 (+ 0.71) to DENV1, 28.21 (+ 0.31) to DENV2, 25.79 (+ 0.63) to DENV3, and 26.92 (+ 0.48) to DENV4.
DiscussionDue to the current epidemiological situation in Brazil, with introduction of Chikungunya and Zika, since 2014 and 2015, respectively, and the continuous challenge of dengue, the ZDC molecular assay (Bio-Manguinhos) proved to be a useful tool for accurate detection, discrimination and epidemiological surveillance of these endemic diseases with similar clinical manifestation.
The performance of the ZDC molecular assay (Bio-Manguinhos) was verified using virus culture, international/commercial panels and synthetic RNA, each of them having results in different units makes a comparison difficult, especially because of the non-comparable unit pfu/mL. However, the virus culture from ZIKV and CHIKV came from LAVIMOAN, the same laboratory that provided the material during the development and validation of the ZDC molecular assay (Bio-Manguinhos). So, the same quantification protocol was used. Comparing the results of this study with the product insert, the same sensitivity is reported to CHIKV and an improved sensitivity was found to ZIKV.
One limitation of this study was the lack of synthetic RNA from DENV. Unfortunately, the material ordered arrived degraded to our laboratory. When the commercial panel was used, DENV results have shown an improved sensitivity. Since the product registration at ANVISA until now, some improvements have been made to the product (data not shown). This is part of a strategy of continuous improvement and it will culminate with a new version of the ZDC molecular assay (Bio-Manguinhos) which will soon incorporate the typing of DENV in the assay.
It is worth highlighting that the IC from ZDC molecular assay (Bio-Manguinhos) added to each sample before the nucleic acid extraction step is fundamental to control the effectiveness of the assay, since the extraction step is the user's responsibility.
The clinical performance of ZDC molecular assay (Bio-Manguinhos) showed 100% sensitivity and 100% specificity to detect and discriminate ZIKV, CHIKV and DENV from clinical plasma samples. Clinical similarities of these arboviruses make their diagnoses a challenge for clinicians and health authorities. The co-circulation and possibility of co-infections reinforce the importance of a diagnostic tool that includes detection of all three viruses and differentiates among them. The ZDC molecular assay (Bio-Manguinhos) results were highly reproducible (100%) with no cross-reaction when testing a panel of other infectious agents.
In conclusion, this broad evaluation demonstrated the capacity of ZDC molecular assay (Bio-Manguinhos) to detect ZIKV, DENV and CHIKV RNA, and discriminate among them, in plasma specimen, with high sensitivity and specificity. The ZDC molecular assay (Bio-Manguinhos) is an accurate and reliable tool to monitor Zika, dengue and chikungunya infections in countries like Brazil with simultaneous circulation of the three viruses.
Ethical approvalThis study was performed in accordance with The Declaration of Helsinki and the Nuremberg Code, complying with the rules for medical research involving humans subjects of the National Health Council and ethical principles. Ethical approval from Institutional Boards were not required for this study because all molecular testing were conducted in addition to the official diagnostic proceedings carried out at Central Laboratories (LACENs) in Brazil.
FundingThis work was supported by Oswaldo Cruz Foundation – FIOCRUZ.
Conflicts of interestThe authors declare no conflicts of interest.