Toxigenic strains of Clostridium difficile may be disseminating. Here we prospectively screened patients with nosocomial diarrhoea in two hospitals in Brazil. To identify C. difficile polymerase chain reaction ribotypes 027/078 strains, we used high resolution melting and multiplex polymerase chain reaction. Among 116 screened patients, 11 were positive for C. difficile. The polymerase chain reaction ribotypes 027/078 strains were not identified in this study.
Clostridium difficile is the main infectious agent of nosocomial diarrhoea, which may result in significant morbimortality.1 Recently, toxigenic strains of C. difficile have emerged, sometimes causing outbreaks.2 Most of these infections are caused by the polymerase chain reaction (PCR) ribotypes 027 and 078 also referred to as BI/NAP1 and BK/NAP7, respectively. Very limited data are available on the frequency of toxigenic strains in Latin America. A study from Costa Rica revealed that 54% (20/37) isolates of C. difficile were identified as C. difficile PCR ribotype 027.3 No other study has documented toxigenic C. difficile strains in Latin America. Here we investigated whether strains 027/078 were present in cases of nosocomial diarrhoea in a cohort study in Brazil.
This was a multicenter prospective observational study, conducted in two hospitals in Porto Alegre, Brazil.4 Hospital 1 is a 65-bed transplant hospital; Hospital 2 is a 800-bed tertiary hospital, including 40 intensive care unit beds. Inpatient adults (>18 years-old) passing ≥3 liquid stools over a 24h period were included. Participation in the study was conditioned to signing an informed consent (ISCMPA Ethics Committee approval – 304/010).
Faecal samples were inoculated on C. difficile agar (CLO) selective medium (bioMérieux, France) and incubated anaerobically for 48h at 37°C. DNA was extracted from bacterial colonies using QIAamp DNA Stool Mini Kit, Qiagen. Molecular characterization of strains was performed by conventional multiplex PCR (mPCR) test, as previously published.5 mPCRs were adjusted to a final volume of 25μL, containing the following: 2.5μL of 10× PCR buffer (200mM Tris-HCl [pH 8.4], 500mM KCl), 2mM MgCl2, 260mM of each deoxyribonucleotide, 1.5U of Platinum Taq DNA Polymerase (Invitrogen), primers (same primers and concentrations as used in Persson et al.6), and 5μL of genomic DNA. Positive controls used in this reaction were obtained from the Statens Serum Institut – Denmark (C. difficile 027 – F62 and C. difficile 078 – M16). Milli-Q® water was used as a negative control. All mPCRs were performed in a Veriti thermal cycler (Life Technologies). Cycling conditions were: 10min at 94°C, followed by 35 cycles of 50s at 94°C, 40s at 54°C, 50s at 72°C, and a final extension of 3min at 72°C.
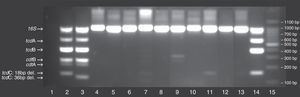
Amplicons sizes were checked by 3.0% (w/v) agarose gel-electrophoresis stained with ethidium bromide. The amplicons tcdC (126 and 144bp, resulting from internal deletions of sizes 39 and 18bp, respectively), as well as the amplicons 16S rDNA, tcdA, tcdB and tcdA/tcdB, were used as standards in each reaction.
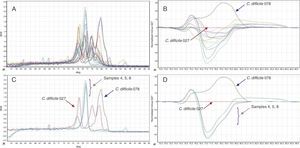
Besides the molecular characterization of C. difficile by mPCR, we optimized a qPCR using high resolution melting (HRM) analysis, adapting the protocol described by Grando et al.7 Type-it HRM PCR (Qiagen) was used to prepare the mix. The PCR mixtures were optimized for a volume of 50μL, containing 25μL of 2× HRM PCR Master Mix (Qiagen), 0.5μM of both forward and reverse primers, 15μL Milli-Q water and 5μL of the genomic template DNA (or Milli-Q water as no template control). Thermocycling conditions and curve normalization were performed according the protocol proposed by Grando et al.7 After normalizing the curves, results were interpreted by subtracting the melting-curve shapes generated by the standard (C. difficile 027 reference strain) from the curves generated for the other isolates and standards.
From the 116 patients included in the study, 11 had positive cultures for C. difficile, and these were evaluated for the presence of hypervirulent characteristics. Among these, 10 clinical strains were non-toxigenic while one strain harboured tcdA and tcdB. When compared to controls, none of the isolates had toxin gene profiles similar to ribotypes 027/078 (Fig. 1). These results were confirmed by HRM, in which none of the isolates showed peaks compatible with PCR ribotypes 027/078 (Fig. 2).
Typing of C. difficile using multiplex conventional polymerase chain reaction (PCR) on 11 clinical isolates. Lane 1: no template control; Lane 2: C. difficile 027 (positive control strain); Lane 3, C. difficile 078 (positive control strain); Lane 15: 100bp DNA ladder; Additional lanes: clinical samples numbered according to their identification in the study; The position of the different C. difficile gene toxins (i.e., tcdA, tcdB, cdtA, cdtB, tcdC) as well as 16S rDNA, internal control are marked on the gel, including the deletions for tcdC.
(A) Melt peak curves on the HRM analysis for the two positive control strains (C. difficile 027/078) and the 11 clinical isolates included in this study. (B) Two positive control strains as well as clinical isolates 4, 5 and 8. (C) Difference plotting of the HMR curves for C. difficile 078 positive control strain, in addition to 11 clinical isolates; C. difficile 027 is used as reference in the graphic. (D) Two control strains (027/078) and clinical isolates 4, 5 and 8.
Molecular typing of C. difficile is still not widely used in tertiary hospitals in Brazil, despite the known emergence of toxigenic C. difficile strains. Proper typing of C. difficile may be of relevance for infection control purposes. Several methods for the differentiation of C. difficile isolates are currently available, including PCR-ribotyping, restriction endonuclease analysis, and pulsed-field gel electrophoresis.8 Performing more than one typing method could potentially increase the discriminatory power of a single typing method, even though few laboratories follow this practice.2 The mPCR is a rapid and specific method for screening toxin genes of C. difficile. This is a simple and relatively cheap method that allows for the presumptive typing of C. difficile strains.6 On the other hand, HRM analysis of PCR products can identify the different genotypic variations among different bacteria. Despite being a more expensive method, HRM offers quicker results and has demonstrated a discriminatory power as high as 0.928.7
In this study, no patient was found to be infected by C. difficile PCR ribotype 027/078 strains. Similar findings were described in a previous study in Brazil, in which ribotyping was performed.9 So far, these strains have not been documented in Brazil. However, considering that molecular typing technologies and anaerobic cultures are still limited in the country, underdetection of C. difficile ribotypes 027/078 strains is also a possibility.9 Even though PCR ribotype 027/078 strains were not detected in this study, our analysis revealed the presence of nine different HRM C. difficile profiles (data not shown).
This study has some limitations. Unfortunately it was not possible to classify the samples that were negative for PCR ribotype 027/078, according to the nomenclature proposed by Stubbs et al.10 Also, despite being a prospective multicenter study, only a small number of isolates tested positive for C. difficile (11/116). Increasing sample size could potentially result in the identification of hypervirulent C. difficile strains. As all isolates showed different peaks in comparison to controls, the discriminatory power of the methods for the detection of C. difficile 027/078 could not be determined.
In conclusion, this study did not detect the presence of C. difficile 027/078 hypervirulent strains in a prospective cohort of patients with nosocomial diarrhoea in Brazil. The importance of implementing molecular typing methods for C. difficile in a continental country such as Brazil deserves additional attention.
Conflicts of interestThe authors declare no conflicts of interest.