The spread of pandemic Staphylococcus aureus clones, mainly methicillin-resistant S. aureus (MRSA), must be kept under surveillance to assemble an accurate, local epidemiological analysis. In Ecuador, the prevalence of the USA300 Latin American variant clone (USA300-LV) is well known; however, there is little information about other circulating clones. The aim of this work was to identify the sequence types (ST) using a Multiple-Locus Variable number tandem repeat Analysis 14-locus genotyping approach. We analyzed 132 S. aureus strains that were recovered from 2005 to 2013 and isolated in several clinical settings in Quito, Ecuador. MRSA isolates composed 46.97% (62/132) of the study population. Within MRSA, 37 isolates were related to the USA300-LV clone (ST8-MRSA-IV, Panton-Valentine Leukocidin [PVL] +) and 10 were related to the Brazilian clone (ST239-MRSA-III, PVL−). Additionally, two isolates (ST5-MRSA-II, PVL−) were related to the New York/Japan clone. One isolate was related to the Pediatric clone (ST5-MRSA-IV, PVL−), one isolate (ST45-MRSA-II, PVL−) was related to the USA600 clone, and one (ST22-MRSA-IV, PVL−) was related to the epidemic UK-EMRSA-15 clone. Moreover, the most prevalent MSSA sequence types were ST8 (11 isolates), ST45 (8 isolates), ST30 (8 isolates), ST5 (7 isolates) and ST22 (6 isolates). Additionally, we found one isolate that was related to the livestock associated S. aureus clone ST398. We conclude that in addition to the high prevalence of clone LV-ST8-MRSA-IV, other epidemic clones are circulating in Quito, such as the Brazilian, Pediatric and New York/Japan clones. The USA600 and UK-EMRSA-15 clones, which were not previously described in Ecuador, were also found. Moreover, we found evidence of the presence of the livestock associated clone ST398 in a hospital environment.
Staphylococcus aureus is one of the most prevalent human pathogens causing a wide range of infectious diseases. Since the introduction of methicillin as a therapeutic option, the emergence and subsequent spread of MRSA has become a worldwide concern. The acquisition of the staphylococcal cassette chromosome mec (SCCmec), which includes the gene mecA of methicillin resistance, was the primary event leading to the appearance of MRSA isolates.1–3 Evidence suggests that this event occurred several times around the world, in different genetic backgrounds, leading to the wide biodiversity of S. aureus strains. Some of these clones are predominant in specific geographic regions, while others have become pandemics.4,5 Three groups of MRSA isolates can be recognized: hospital associated MRSA (HA-MRSA), community associated MRSA (CA-MRSA), and livestock associated MRSA (LA-MRSA) strains.6 Moreover, the epidemiological behavior of methicillin-susceptible S. aureus (MSSA) remains an important area of study in clinical settings around the world.5
First reported in the 1960s, MRSA strains were initially confined to the hospital environment.6 HA-MRSA encompasses diverse epidemic and pandemic clones, which vary with geography. For instance in the USA, the most prevalent HA-MRSA clones are ST5-MRSA-II (USA100/NY-Japan) and ST5-MRSA-IV (USA800/Pediatric), while in the UK the most prevalent HA-MRSA clone is the ST22-MRSA-IV (UK-EMRSA-15) clone. In South America, the most disseminated HA-MRSA clone is the ST239-MRSA-III (Brazilian Epidemic Clone [BEC]).3,7–9 Even though, the Cordobes/Chilean (ST5-MRSA-I) clone was found as the most prevalent in countries like Chile and Argentina.3,8–10 The ST5-MRSA-II and a variant of the ST239-MRSA-III are the most prevalent clones in Asia.8
In the1990s, infections with MRSA strains in patients without risk factors began to be reported.11 These CA-MRSA strains had emerged almost simultaneously around the world, by independent events of SCCmec acquisition, constituting different epidemic clones.12 In Europe for instance, the ST80-MRSA-IV strains (European clone) are widely spread, and in East Asia the ST59-MRSA-IV clone remains common.13,14 In North America, the most prevalent is the epidemic USA300 clone (ST8-MRSA-IV), which has extensively replaced previous clones, such as USA100 and other HA-MRSA clones, mainly in USA.11,15 The USA300 clone is actually considered pandemic, as it has been isolated repeatedly in every continent.12 Recently, a Latin American native CA-MRSA clone, closely related to the pandemic USA300, was described as prevalent in the northern South America region, in both hospital settings and community environments.16 This ST8-MRSA-IV clone, USA300-LV, has been found to carry Panton Valentine Leukocidin (PVL) haplotype R.17 Since it was first reported in 2006, studies have demonstrated that this clone has fully replaced the previously established Chilean/Cordobes clone in northern South America.18
Another group of S. aureus, the livestock associated S. aureus, has become a worldwide threat associated with the handling of farm animals.19 The main clone associated with livestock includes strains belonging to the ST398 lineage, and both MRSA and MSSA strains.20,21 Since its primary identification in the Netherlands in 2003 in domestic pigs, this clone has been reported in most of European countries, North America and Asia, not only in pigs but also in cattle, horses, poultry, and humans.6,20 Initially, the carriage and infection of these strains were associated with direct contact with animal farming as the major risk factor. Currently, increasing reports describe invasive infections with ST398 in patients with no previous livestock environment contact.22
There is little information about the clones of either HA-MRSA or CA-MRSA circulating in Ecuador. In a regional, multicenter study in 2009, we described the clones established in Colombia, Venezuela, Peru, and Ecuador.16 The study focused on CA-MRSA strains, and it provided evidence of the extensive presence of the USA300-LV clone in Ecuador. Moreover, we identified a significant presence of the HA-MRSA Brazilian Epidemic Clone.
There are various genotyping methods available to study the epidemiology of S. aureus, each one with different advantages and disadvantages, as well as different resolving power.8 Multiple-Locus Variable number tandem repeat Analysis (MLVA) represents a high-throughput genotyping method characterized by its low cost and high resolution.23,24 To identify the clones of S. aureus circulating in Ecuador, we applied an MLVA typing approach to a heterogeneous strain collection including both MRSA and MSSA isolates.
MethodsBacterial study population and antimicrobial susceptibility profileA total of 132 S. aureus isolates, from different populations, were studied. One set (population 1, P1) consisted of 80 isolates consecutively recovered between 2010 and 2013 from bloodstream infections of patients from three hospitals in Quito, Ecuador. Another set (population 2, P2) contained 32 non-consecutive isolates recovered between 2011 and 2013 from different clinical infections from 11 different clinical settings. Lastly, a third set (population 3, P3) consisted of 20 isolates recovered from different clinical origins between 2005 and 2007. These isolates were randomly selected from a MRSA archived collection belonging to a hospital in Quito. The purpose of analyzing different sets of isolates was to detect a wider variety of local clones other than USA-300-LV.
Antimicrobial susceptibility profiling was performed on all isolates. Minimum inhibitory concentrations (MICs) were obtained for oxacillin, gentamicin, ciprofloxacin, erythromycin, clindamycin, linezolid, vancomycin, tetracycline, tigecycline, rifampicin, and trimethoprim-sulfamethoxazole using the Vitek®2 System, according to manufacturer specifications and CLSI delineations. Additionally, we have tested the susceptibility to cefoxitin (data not shown), in order to rule out the presence of mecC.25
Detection of mecA and Panton-Valentine Leukocidin (PVL) virulence factorsThe presence of the gene mecA, and the lukS-PV and lukF-PV genes, encoding the subunits of PVL, were detected using a PCR-multiplex with specific primers described elsewhere.26,27 Additionally, the PCR-multiplex amplified the S. aureus species specific gene nuc.28 Briefly, one reaction mix contained 5ng of DNA, 12.5μL of GoTaq® Green Master Mix (1× Green GoTaq® Reaction Buffer [pH 8.5], 200μM of each dNTP and 1.5mM MgCl2, Promega) and 0.4μM of each primer in a final volume of 25μL. The PCR program included 35 cycles of 30s of denaturation at 94°C, 30s of annealing at 62°C and 1min of extension at 72°C. The PCR products of PVL were sequenced in selected isolates to identify the haplotype.29
Screening for the SCCmec typeWe carried out the SCCmec typing using the Chen, et al. (2009) scheme, with modifications. Conventional single PCR was performed using primers specific to genes of mec and ccr complexes.30 The PCR reactions contained 5ng of DNA, 12.5μL of GoTaq® Green Master Mix and 0.4μM of each primer in a final volume of 25μL. The PCR program included 35 cycles of 30s of denaturation at 95°C, 30s of annealing at 55°C and 30s of extension at 70°C. The PCR products were visualized in 3% UltraPure™ Agarose-1000 electrophoresis gels (Promega).
Multiple-Locus Variable number of tandem repeat Analysis (MLVA) typingWe used specific primers to independently amplify a 14 VNTR panel known as MLVA 14Orsay.23,31 PCR reactions were performed in 25μL reactions containing 5ng of DNA, 12.5μL of GoTaq® Green Master Mix and 0.2μM of each primer. The PCR program consisted of 25 cycles of denaturation at 94°C for 30s, annealing at 60°C for 30s and extension at 72°C for 30s. The PCR products were resolved in 2% agarose electrophoresis gels. The exact copy number of the repetitive unit in each locus was calculated as previously described.23 Four strains with a previously known copy number of repetitive units in each VNTR were use as controls and were included in every run. The sequence type of the four MLVA control strains are ST5, ST8, ST30, and ST45, which were confirmed by MLST as described elsewhere.32 We identified the ST related to the 132 isolates by querying the MLVA genotypes of each isolate in the MLVAbank for Bacterial Genotyping (http://mlva.u-psud.fr/ [S. aureus database]). Simpson's diversity index was applied.33 A UPGMA dendrogram was constructed using the categorical distance coefficient, applying a cut-off of 45% similarity.23 A minimum spanning tree (MST) was built using the PHYLOViZ 1.1 software.
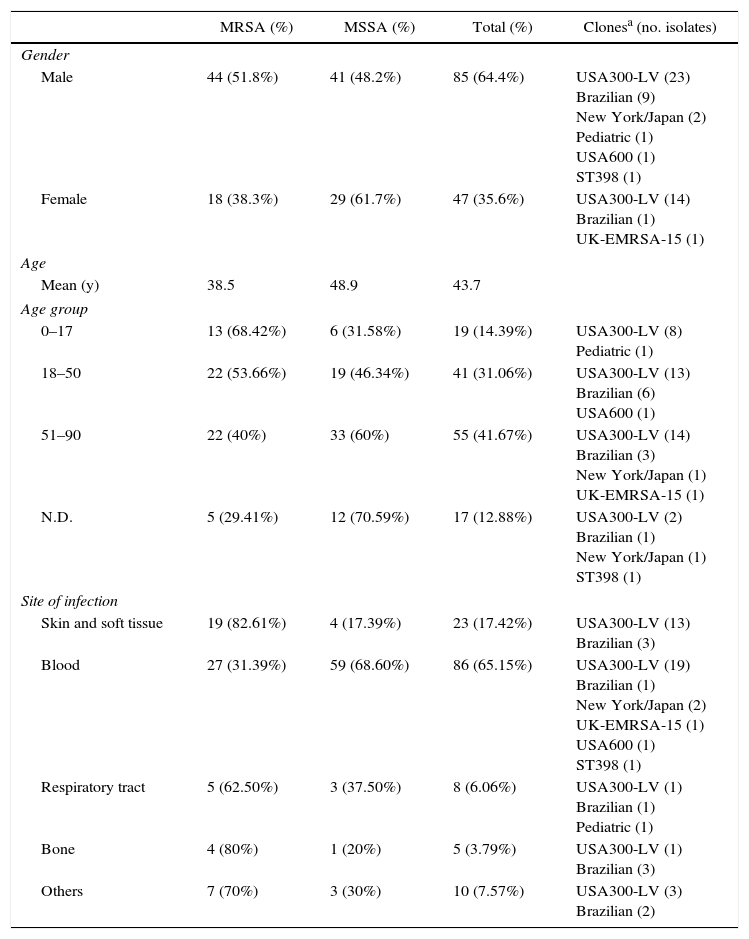
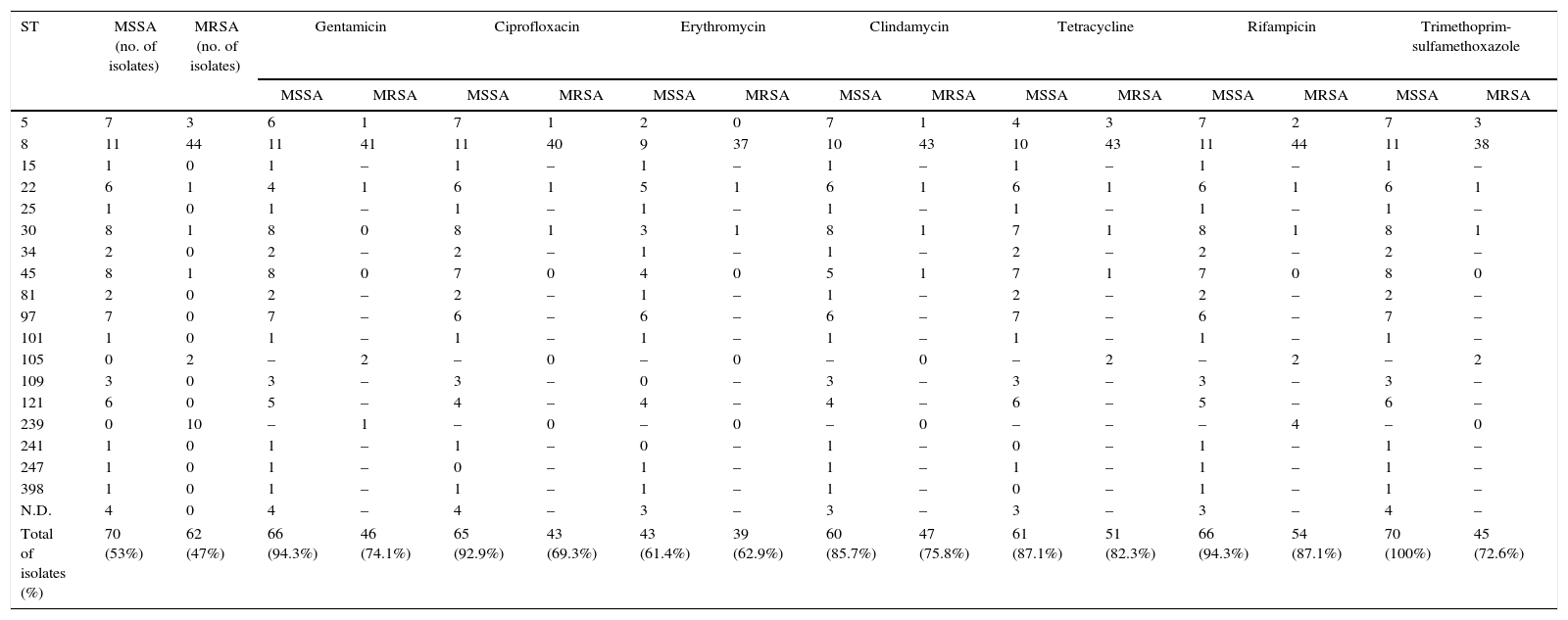
ResultsIsolate population and antimicrobial profileThe origin, sex and age of the patients from which the 132 isolates were taken are summarized in Table 1. All isolates were susceptible to vancomycin, linezolid, and tigecycline. Sixty-two (47%) isolates were MRSA. Of these, 74.1% were susceptible to gentamicin, 69.3% to ciprofloxacin, 62.9% to erythromycin, 75.8% to clindamycin, 82.3% to tetracycline, 87.1% to rifampicin, and 72.6% to trimethoprim-sulfamethoxazole. On the other hand, 70 (53%) isolates were MSSA. Of these, 94.3% were susceptible to gentamicin, 92.9% to ciprofloxacin, 61.4% to erythromycin, 85.7% to clindamycin, 87.1% to tetracycline, 94.3% to rifampicin, and 100% to trimethoprim-sulfamethoxazole (Table 2). All MRSA isolates were cefoxitin resistant and all MSSA isolates were cefoxitin susceptible.
Patient characteristics from whom MRSA and MSSA have been recovered.
| MRSA (%) | MSSA (%) | Total (%) | Clonesa (no. isolates) | |
|---|---|---|---|---|
| Gender | ||||
| Male | 44 (51.8%) | 41 (48.2%) | 85 (64.4%) | USA300-LV (23) Brazilian (9) New York/Japan (2) Pediatric (1) USA600 (1) ST398 (1) |
| Female | 18 (38.3%) | 29 (61.7%) | 47 (35.6%) | USA300-LV (14) Brazilian (1) UK-EMRSA-15 (1) |
| Age | ||||
| Mean (y) | 38.5 | 48.9 | 43.7 | |
| Age group | ||||
| 0–17 | 13 (68.42%) | 6 (31.58%) | 19 (14.39%) | USA300-LV (8) Pediatric (1) |
| 18–50 | 22 (53.66%) | 19 (46.34%) | 41 (31.06%) | USA300-LV (13) Brazilian (6) USA600 (1) |
| 51–90 | 22 (40%) | 33 (60%) | 55 (41.67%) | USA300-LV (14) Brazilian (3) New York/Japan (1) UK-EMRSA-15 (1) |
| N.D. | 5 (29.41%) | 12 (70.59%) | 17 (12.88%) | USA300-LV (2) Brazilian (1) New York/Japan (1) ST398 (1) |
| Site of infection | ||||
| Skin and soft tissue | 19 (82.61%) | 4 (17.39%) | 23 (17.42%) | USA300-LV (13) Brazilian (3) |
| Blood | 27 (31.39%) | 59 (68.60%) | 86 (65.15%) | USA300-LV (19) Brazilian (1) New York/Japan (2) UK-EMRSA-15 (1) USA600 (1) ST398 (1) |
| Respiratory tract | 5 (62.50%) | 3 (37.50%) | 8 (6.06%) | USA300-LV (1) Brazilian (1) Pediatric (1) |
| Bone | 4 (80%) | 1 (20%) | 5 (3.79%) | USA300-LV (1) Brazilian (3) |
| Others | 7 (70%) | 3 (30%) | 10 (7.57%) | USA300-LV (3) Brazilian (2) |
Number of susceptible isolates according to the sequence type (ST).
| ST | MSSA (no. of isolates) | MRSA (no. of isolates) | Gentamicin | Ciprofloxacin | Erythromycin | Clindamycin | Tetracycline | Rifampicin | Trimethoprim-sulfamethoxazole | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MSSA | MRSA | MSSA | MRSA | MSSA | MRSA | MSSA | MRSA | MSSA | MRSA | MSSA | MRSA | MSSA | MRSA | |||
| 5 | 7 | 3 | 6 | 1 | 7 | 1 | 2 | 0 | 7 | 1 | 4 | 3 | 7 | 2 | 7 | 3 |
| 8 | 11 | 44 | 11 | 41 | 11 | 40 | 9 | 37 | 10 | 43 | 10 | 43 | 11 | 44 | 11 | 38 |
| 15 | 1 | 0 | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – |
| 22 | 6 | 1 | 4 | 1 | 6 | 1 | 5 | 1 | 6 | 1 | 6 | 1 | 6 | 1 | 6 | 1 |
| 25 | 1 | 0 | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – |
| 30 | 8 | 1 | 8 | 0 | 8 | 1 | 3 | 1 | 8 | 1 | 7 | 1 | 8 | 1 | 8 | 1 |
| 34 | 2 | 0 | 2 | – | 2 | – | 1 | – | 1 | – | 2 | – | 2 | – | 2 | – |
| 45 | 8 | 1 | 8 | 0 | 7 | 0 | 4 | 0 | 5 | 1 | 7 | 1 | 7 | 0 | 8 | 0 |
| 81 | 2 | 0 | 2 | – | 2 | – | 1 | – | 1 | – | 2 | – | 2 | – | 2 | – |
| 97 | 7 | 0 | 7 | – | 6 | – | 6 | – | 6 | – | 7 | – | 6 | – | 7 | – |
| 101 | 1 | 0 | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – |
| 105 | 0 | 2 | – | 2 | – | 0 | – | 0 | – | 0 | – | 2 | – | 2 | – | 2 |
| 109 | 3 | 0 | 3 | – | 3 | – | 0 | – | 3 | – | 3 | – | 3 | – | 3 | – |
| 121 | 6 | 0 | 5 | – | 4 | – | 4 | – | 4 | – | 6 | – | 5 | – | 6 | – |
| 239 | 0 | 10 | – | 1 | – | 0 | – | 0 | – | 0 | – | – | – | 4 | – | 0 |
| 241 | 1 | 0 | 1 | – | 1 | – | 0 | – | 1 | – | 0 | – | 1 | – | 1 | – |
| 247 | 1 | 0 | 1 | – | 0 | – | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – |
| 398 | 1 | 0 | 1 | – | 1 | – | 1 | – | 1 | – | 0 | – | 1 | – | 1 | – |
| N.D. | 4 | 0 | 4 | – | 4 | – | 3 | – | 3 | – | 3 | – | 3 | – | 4 | – |
| Total of isolates (%) | 70 (53%) | 62 (47%) | 66 (94.3%) | 46 (74.1%) | 65 (92.9%) | 43 (69.3%) | 43 (61.4%) | 39 (62.9%) | 60 (85.7%) | 47 (75.8%) | 61 (87.1%) | 51 (82.3%) | 66 (94.3%) | 54 (87.1%) | 70 (100%) | 45 (72.6%) |
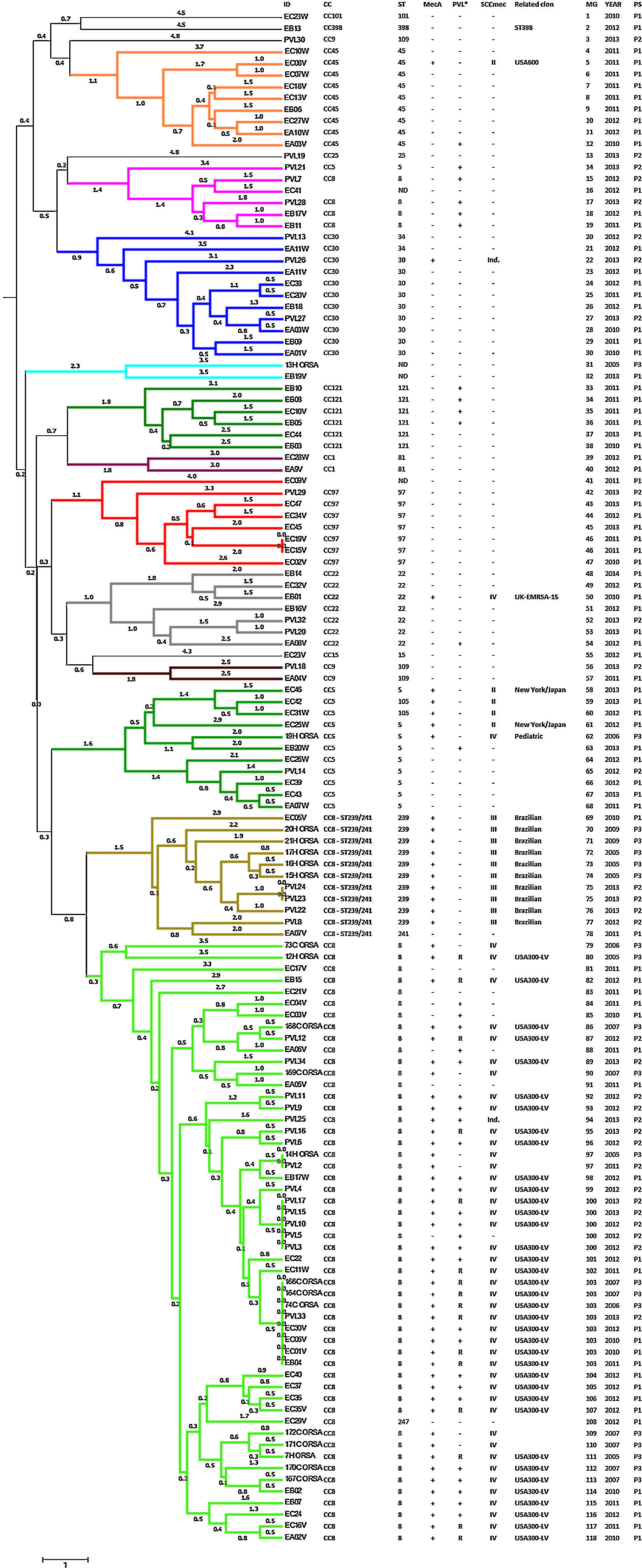
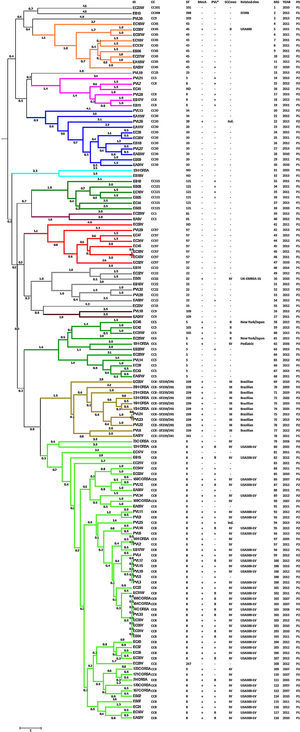
The presence of the mecA methicillin resistant gene was detected in all isolates identified as MRSA using the Vitek®2 System (46.97%). The PVL virulence factor was detected in 56 isolates (42.42%): 67.86% of MRSA isolates and 32.14% of MSSA isolates (Fig. 1). SCCmec type II was identified in five isolates, SCCmec type III in 10 isolates, and SCCmec type IV was detected in 45 isolates. The typing scheme failed to identify the SCCmec type in two isolates. Querying the MLVA genotypes against the MLVA bank for Bacterial Genotyping database we identified 18 different sequence types (ST). Fifty-five isolates (41.67%; 44 MRSA and 11 MSSA) belonged to ST8. All of the ST8 MRSA isolates presented the SCCmec type IV genotype, except one cataloged as indeterminate. The ST8-MRSA-IV-PVL positive profile (USA300-LV related) was detected in 37 isolates. PVL haplotype was analyzed in 16 USA300-LV related isolates and all were identified as R. The ST8-MRSA-PVL negative profile was detected in six isolates, whereas ST8-MSSA-PVL positive profile was detected in nine isolates. Ten isolates were identified as ST239, all of which presented the SCCmec type III and were PVL negative (Brazilian Epidemic Clone related). Ten and two isolates belonged to ST5 and ST105 (MLST Single Locus Variant [SLV] of ST5), respectively. Three of the ST5 isolates were MRSA; two of these were SCCmec type II, related to the New York/Japan clone, and one isolate was SCCmec type IV, related to the Pediatric clone. The two ST105 isolates were SCCmec type II. Only two ST5 MSSA strains were PVL positive. Other representative STs found here were ST45 and ST30 with nine isolates each. One ST45 isolate had the USA600 profile (ST45-MRSA-II, PVL negative) and only one ST30 was MRSA with an indeterminate SCCmec type. Seven isolates were ST22, one of which had the UK-EMRSA-15 profile (ST22-MRSA-IV, PVL negative). Additionally, seven and six isolates were ST97 and ST121, respectively, all of them were MSSA. We also identify one MSSA isolate related to the livestock associated ST398 clone. Four isolates could not be genotyped as a specific ST (Fig. 1).
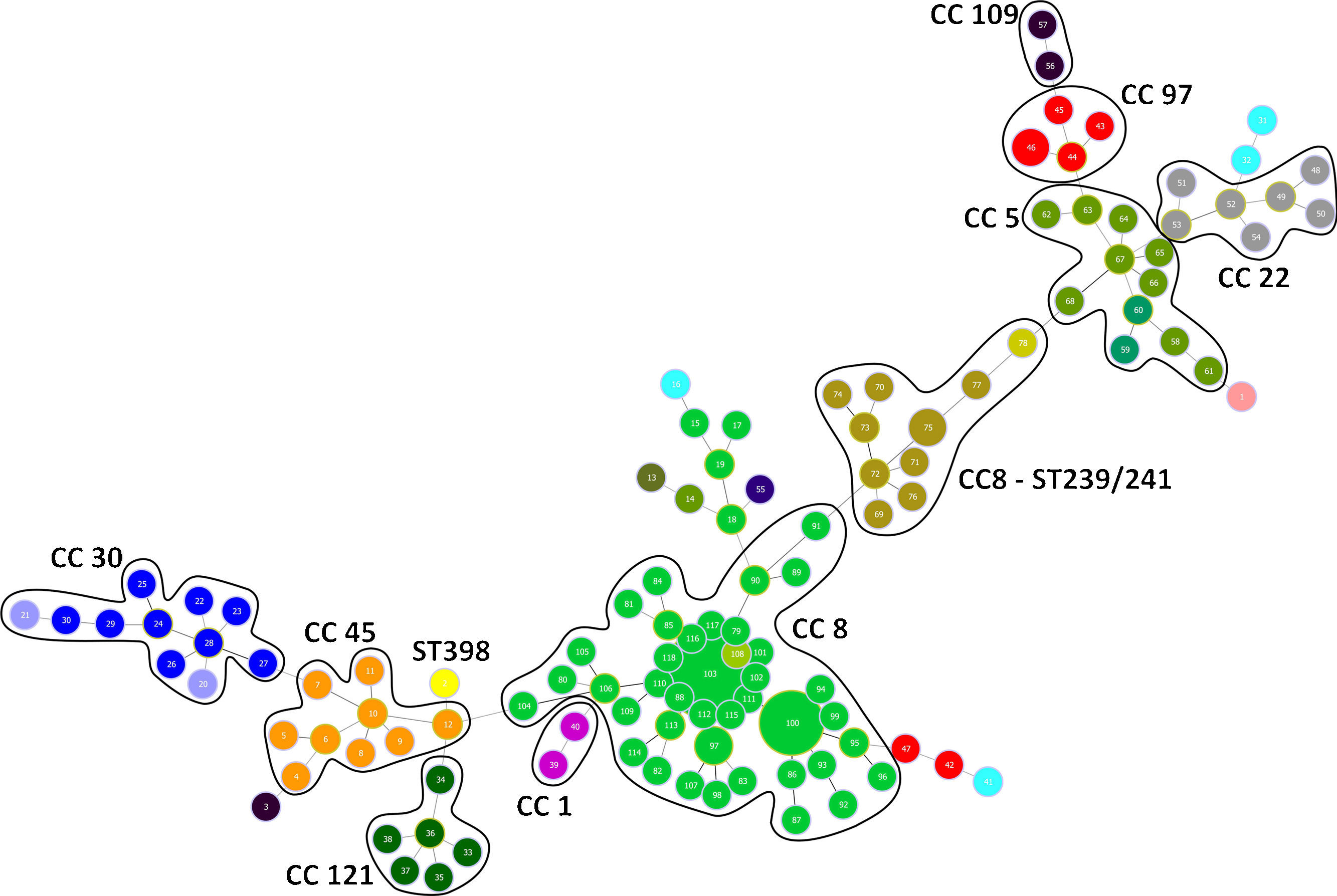
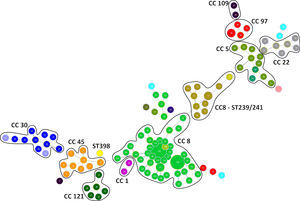
Genotyping analysis of MRSA and MSSA isolatesAll 132 isolates were resolved into 118 MLVA genotypes with an overall Simpson's diversity index of 0.99. The 62 MRSA isolates and the 70 MSSA isolates belonged to 50 and 69 MLVA genotypes, respectively. From all MLVA genotypes, 12 clusters were identified using a 45% cut-off value in a UPGMA tree, which includes 127 (96.21%) isolates (Fig. 1). These clusters were included among the major Clonal Complexes (CC) identified in the MST analysis (Fig. 2). The largest cluster was CC8, with 52 (39.39%) isolates, including 37 USA300-LV related isolates. The clusters CC5, CC8 – ST239/241 and CC30 had 11 isolates each, including two New York/Japan related isolates (CC5), one Pediatric related isolate (CC5) and 10 Brazilian related isolates. The CC1 (two isolates), CC22 (seven isolates), CC45 (nine isolates), CC97 (eight isolates), CC109 (two isolates), and CC121 (six isolates) clusters accounted for 25.76% of the studied collection, including one USA600 related isolate (CC45) and one UK-MRSA-15 related isolate (CC22). One cluster had a heterogeneous composition with isolates belonging to ST8 (four isolates), ST5 (one isolate), and an unspecified ST (one isolate). Furthermore, one cluster was composed of two unspecific STs. The remaining five isolates belonged to ST101, ST109, ST25, ST15, and the livestock associated ST398.
DiscussionEpidemic clones of S. aureus have successfully spread worldwide. Some of these clones have reached high prevalence at the local level (Cordobes/Chilean clone), and others have become the most prevalent circulating clones at a regional level (USA300, LA-MRSA ST398 or UK-MRSA-15 clones).8 In Latin America, native clones such as the Brazilian, Cordobes/Chilean and Pediatric clones, and “imported” clones such as the New York/Japan and USA300 clones have been described.9 In Ecuador, only two clones had been described; the predominant USA300-LV clone, and the Brazilian clone, which has lower prevalence.16,18 In this study we identified additional clones using the MLVA genotyping tool. This molecular tool is a high-throughput typing method, characterized by its low cost and high discriminatory power. MLVA has proven to be a reliable typing method with high concordance with other typing methods such as PFGE and MLST.16,23,24,34
We found a high number of isolates related to the USA300 clone (ST8-MRSA-IV, PVL positive), confirming the predominance of this clone in Ecuador.16 These isolates form a defined cluster in both UPGMA and MST analysis. The related clone was originally identified in the USA in 2000 after causing skin and soft issue infections in football players in Pennsylvania and prisoners in Missouri. At that time, USA300 was the most prevalent CA-MRSA clone isolated.15 We identified USA300 related isolates in P1, P2 and P3, demonstrating that this clone was already circulating in Ecuador as early as 2005. The first reports of this clone in the northern South America region came from Colombia, also in 2005.35 The ST8-MRSA-IV, PVL negative isolates found in this study could be related to Lyon Clone/UK-EMRSA-2, frequently isolated in France; UK-EMRSA-6, or USA500, identified in the USA, Australia, the UK, Ireland, and Sub-Saharan Africa.6 It is an interesting highlight that the presence of the PVL virulence factor was linked to ST8 related isolates; only eight isolates from different STs were PVL positive. Moreover, the sequence of the lukS-PV and lukF-PV genes of the 16 ST8-MRSA-IV-PVL positive isolates analyzed here identified the haplotype R in all of them. The PVL haplotype R is strongly related to MRSA strains especially those of ST8 lineage.17,29 The ST8 related isolates represent 70.97% of the MRSA isolates found in this study, underscoring the success of this ST in the region.11,12,16,18,35,36 Moreover, many of the isolates related to ST8 showed susceptibility to all antibiotics tested in this work. This extensive susceptibility profile is described as characteristic to USA300 clone strains.15 Only 10 of the ST8 isolates were resistant to erythromycin, gentamicin, ciprofloxacin, or trimethoprim-sulfamethoxazole.
Previously, we had identified the Brazilian clone as the most prevalent HA-MRSA clone in Ecuador.16 Here, we detected 10 isolates related to this clone (ST239-MRSA-III), the second most prevalent ST found in this study. The ST239 clonal group, recognized as one of the oldest pandemic MRSA strains, is the result of a chromosomal recombination event involving parental ST8 and ST30 strains.6,37,38 The ST239 clonal group has been reported worldwide and it is divided into three clades, I, II and III.37 The Brazilian clone, belonging to clade III, was reported in early 1990s in Brazil and is now identified in virtually the entire region.6,9,37,39 Here, we identified isolates related to the ST239 clone in samples from 2005, 2009, 2010, 2012, and 2013. The random choice of samples did not allow us to estimate the real prevalence of ST239, but this analysis of samples taken over several years did give us an idea of the timing of the presence of this clone in Ecuador. Additionally, these ST239 isolates have a typical resistance profile reported for this clone: ciprofloxacin, erythromycin, clindamycin, and trimethoprim-sulfamethoxazole resistance.9
There is no record of isolates related to CC5 in Ecuador. In this study, we identified three isolates related to CC5 MRSA pandemic clones: two isolates related to the New York/Japan clone (ST5-MRSA-II) and one isolate related to the Pediatric clone (ST5-MRSA-IV). The New York/Japan clone was described in the mid-1990s as the MRSA strain most prevalent in New York metropolitan hospitals. Later studies demonstrated that this clone was dominant in Japan in the early 1990s, showing the transcontinental spread of the clone.40 The New York/Japan clone has been described in the USA, the UK, Portugal, Germany, and other European countries. It has also been described in Asia, specifically China and Taiwan, and in Australia.6 In South America, this clone was detected in Brazil,9 but not in neighboring countries, such as Colombia and Peru. These two isolates, related to the New York/Japan clone, were recovered from P1, in 2012 and 2013 at the same hospital, but their sporadic occurrence suggests that these two ST5-MRSA-II isolates are not related. On the other hand, the one isolate related to the Pediatric clone (ST5-MRSA-IV) was detected in P3, from 2006, suggesting the displacement of this clone.
The UK-EMRSA-15 clone (ST22-MRSA-IV) is the most prevalent HA-MRSA clone in some regions of Europe (UK, Portugal and Ireland) and Singapore.1,6,41 In South America the ST22-MRSA-IV profile was only recorded from one isolate from Venezuela.16 We found one isolate with the UK-EMRSA-15 profile, probably representing the second record of this clone in South America. Another important clone found in this study was ST45-MRSA-II, known as the USA600 clone. This clone appears to be largely restricted to North America, with sporadic reports from Hong Kong and Australia.6 In South America it has been recorded in Uruguay.9
The principal MSSA isolates identified were ST5, ST8, ST22, ST30, ST45, and ST97. Some MSSA strains are the ancestral strains of epidemic MRSA clones, such as ST30 MSSA, which evolved into the Southwest Pacific Clone (USA1100).42,43 We found several isolates to have the same ST background as MRSA epidemic clones, including ST8-MSSA, ST5-MSSA, and ST22-MSSA. Because numerous studies have demonstrated that some pandemic MRSA clones could have been originated through intermittent events of acquisition and loss of SCCmec, surveillance of MSSA isolates is important.5,42
Only one isolate each of ST5-MRSA-IV, ST22-MRSA-IV and ST45-MRSA-II was found, and could be cataloged as imported cases or furtive events of SCCmec cassette acquisition.
In South America, LA-MRSA ST398 was reported in Brazil and Peru, associated with cows and pigs, respectively.44,45 In the other hand, LA-MSSA ST398 had been previously described in Colombia as causing infection in a woman without livestock association.46 Here, we found a MSSA isolate, recovered from a blood stream infection (P1), related to the livestock associated clone ST398. MSSA isolates of this clone have been recovered from blood stream infections and suggested as a human adapted sub-clone.22 However, further investigations theorize LA-MRSA/MSSA ST398 clone has human origin as MSSA lineage, which spread into farm animals where it acquired methicillin and tetracycline resistance.20,47 Most of the reports describing ST398 strains isolated from infection in humans without risk factors (livestock contact) are MSSA type and tetracycline susceptible, differing from tetracycline-resistant strains recovered from livestock.21,22,47 Although, our isolate came from a patient without known previous animal contact, the tetracycline resistance suggests an animal origin.
In conclusion, we detected a wide variety of STs in both MRSA and MSSA. We identified the presence of previously undetected clones in Ecuador, as well as confirmed the prevalence of clones that had already been reported in the region, which was made feasible because of the type of sampling that we applied in this study. Our results emphasize the need for a deep analysis of the current epidemic structure of S. aureus to accomplish a more accurate surveillance. Additionally, because of the low cost and powerful discriminatory power of the MLVA 14Orsay genotyping approach, we were able to successfully apply this tool to identify the circulating clones of S. aureus in Quito, Ecuador.
Conflicts of interestThe authors declare no conflicts of interest.
The authors would like to acknowledge the technical assistance of Cecibel Gonzalez. This study was supported by Pontificia Universidad Católica del Ecuador: Project K13106 and Zurita & Zurita Laboratorios: Project MIC-004. Part of this work was presented in 24th ECCMID 2014.