We have evaluated the efficacy of short-interfering RNAs targeting the nucleoprotein gene and also the brain immune response in treated and non-treated infected mice. Mice were inoculated with wild-type virus, classified as dog (hv2) or vampire bat (hv3) variants and both groups were treated or leaved as controls. No difference was observed in the lethality rate between treated and non-treated groups, although clinical evaluation of hv2 infected mice showed differences in the severity of clinical disease (p=0.0006). Evaluation of brain immune response 5 days post-inoculation in treated hv2 group showed no difference among the analyzed genes, whereas after 10 days post-inoculation there was increased expression of 2′,5′-oligoadenylate synthetase 1, tumor necrosis factor alpha, interleukin 12, interferon gamma, and C-X-C motif chemokine 10 associated with higher expression of N gene in the same period (p<0.0001). In hv2 non-treated group only higher interferon beta expression was found at day 5. The observed differences in results of the immune response genes between treated and non-treated groups is not promising as they had neither impact on mortality nor even a reduction in the expression of N gene in siRNA treated animals. This finding suggests that the use of pre-designed siRNA alone may not be useful in rabies treatment.
Rabies virus (RABV) causes acute encephalitis and has a case-fatality rate approaching 100% being considered one of the most deadly existent infectious diseases.1 The survival of a 15-year-old girl from Wisconsin, bitten by a bat that received no vaccination, led physicians worldwide to apply the protocol known as the “Milwaukee Protocol”2 but after 10 years it has been shown to be ineffective. There are at least 26 reported cases in which this protocol was tested without success.3 Therefore, continuous efforts should be made to find some effective treatment for rabies, including new technologies such as RNA interference.
RNA interference (RNAi) is an endogenous mechanism, first described in the late 90s that leads to post-transcriptional gene silencing. It is well conserved in a broad variety of species, including plants and animals.4,5 A short nucleotide sequence (approx. 21–23 nucleotides length), also known as short-interfering RNA (siRNA), associated with the RNA-induced silencing complex (RISC), recognizes and binds to a complementary mRNA, causing its cleavage into smaller fragments and inactivating its expression and, thus, inhibiting protein synthesis.6 The RNAi mechanism plays an important role in cellular defense against viral infections in addition to other important cellular functions, including the mobility of genetic elements and regulation of gene expression during animal development.7 The general potential of this mechanism has stimulated studies of the use of siRNA and microRNA as a therapeutic option for non-infectious8–10 and infectious diseases, including dengue,11,12 respiratory syncytial virus,13 influenza,14 tuberculosis,15 SARS,16 AIDS,10 and herpes simplex type 2.17
Despite the antiviral effect of siRNAs, they are potent activators of the mammalian innate immune system. Synthetic siRNA duplexes can induce high levels of inflammatory cytokines and type I interferons, after systemic administration in mammals and in primary human blood cell cultures.18,19 The production of antiviral agents such as type I interferons, including interferon alpha (IFNα) and interferon beta (IFNβ), is an important immune mechanism against rabies virus infection that occurs soon after the cell infection.20,21
In 2007, Brandão and colleagues published a study in BHK-21 cells showing the efficacy of a novel therapy against rabies virus based on the use of siRNAs designed against the N gene sequence of Pasteur virus (PV). The results demonstrated that cells treated with three different sequences of siRNA had a five-fold drop in the amount of infected cells evaluated by direct immunofluorescence test when compared to controls, with no cytopathogenicity due to the treatment.22 Those same sequences were tested by the same group in vivo and demonstrated reduction in the lethality rate when compared to untreated animals.23
Studies testing siRNAs in vitro and in vivo usually have as targets rabies nucleoprotein (N), glycoprotein (G), and/or polymerase (L) genes; the sequences are delivered by a vector such as adenovirus,24 lentivirus,25 or associated with a liposome.26 siRNAs always inhibit viral replication at some level, however it is difficult to precisely determine their real efficacy and possible application in medical practice. This is because in almost all studies, the siRNAs tested are those designed and checked in experimental infection due to exactly the same RABV strains (usually a laboratory strain) used as templates to design the siRNA sequences.7,22,24,26
This study aimed to test the clinical efficacy of three different sequences of siRNA designed against the RABV N gene in the treatment of mice infected with two different wild strains of RABV, isolated from rabid human patients infected by a dog or by a vampire bat variant. In addition, considering the difference of pathogenicity between dog and bat variants27 and the immune stimulation that siRNA administration can induce, the brain immune response of infected and non-infected animals was evaluated.
Materials and methodsExperimental designTwo groups of 60 C57/BL6 mice each, 4–6 week-old females, S.P.F, were inoculated in the gastrocnemius muscle with 100μL of viral inoculum with same viral titration (LD50 10−6.66/30μL) for variant 2 [dog (hv2)] or variant 3 [vampire bat (hv3)]. Thirty animals were treated intraperitoneally 24h p.i., with a unique dose of a mixture consisting of three siRNA sequences (3.3μM concentration each) designed against the N gene of the PV strain, using lipofectamine as the delivery method22 (Table 1); the other half (n=30) were left untreated, and just received saline intraperitoneally at the same time of the siRNA treated group. A non-inoculated group of animals (n=30) were used as controls for basal immune response and received intramuscular of sterile saline as well as intraperitoneal inoculation. A non-inoculated siRNA treated group (n=30) was included to evaluate possible side effects of the treatment and also the immune stimulation of siRNA. For all groups, 10 animals were observed for 30 days and 10 were euthanized after 5 and 10 days p.i., when whole brains were removed and stored at −80°C until further real-time PCR analyses.
Nucleotide sequences of siRNAs designed against Pasteur virus N gene.22
| siRNA | Duplex sequence |
|---|---|
| RNA124 | Sense 5′GCCUGAGAUUAUCGUGGAG 3′Antisense 5′AUCCACGAUAAUCUCAGGC 3′ |
| RNA750 | Sense 5′GCACAGUUGUCACUGCUUC3′Antisense 5′UAAGCAGUGACAACUGUGC 3′ |
| RNA B | Sense 5′GACAGCUGUUCCUCACUCG 3′Antisense 5′AGAGUGAGGAACAGCUGUC 3′ |
Animals of all groups were weighted and evaluated daily for the onset of rabies clinical signs, such as ruffled fur, hunching back, hypo/hyper excitability, paralysis of one or both hind limb or tetraplegia,28 and for any other abnormality in the case of the non-inoculated groups.
The animal study was approved by the São Paulo State University Ethical Committee (registration number 238/2008), which follows the guidelines established by the COBEA – Brazilian College of Animal Experimentation).
RNA extraction and Real Time-RT-PCR (RT-qPCR)Brain tissue RNAs were extracted with the Invitek® kit and stored at −80°C. The reaction for cDNA synthesis consisted of 1μg of extracted RNA, 1μL of Oligo-DT primer (Invitrogen®) and 1μL of SuperScript II (Invitrogen®) according to the manufacturer's instructions. The RT-qPCR reaction was performed with 2μL of 1/50 diluted cDNA, 1μL of 0.1μg of each primer and Master Mix Syber Green (Promega®) in a final volume of 25μL according to the manufacturer's instructions. Primers for the 18S murine genes were supplied by IDT® and used as housekeeping genes, and primers for the RABV N gene were manufactured as described previously.29 The mouse Quantitect® Primer Assay from Qiagen® was used to evaluate the expression of chemokine C-C motif ligand 2 (CCL2), 2′-5′-oligoadenylate synthetase 1 (OAS1), interleukin 2 (IL2), interleukin 6 (IL6), interleukin 12 (IL12), tumor necrosis factor alpha (TNFα), interferon gamma (IFNγ), interferon beta (IFNβ), C-X-C motif chemokine 10 (CXCL10), cell surface glycoprotein CD200 receptor 1(CD200R) and insulin-like growth factor 1 (IGF-1).
All thermal cycling and detection was performed using an Applied Biosystems StepOne Fast (ABI7500 Fast) thermal cycler employing a thermal profile of 40 cycles of 50°C for 20s, 95°C for 10min, 95°C for 15s and 60°C for 1min.
Data analysisCox proportional hazards were used to estimate lethality rate and hazard ratios (HR) between groups. Kruskal–Wallis with p<0.05 as the significance level was chosen for evaluation of gene expression of cytokines/chemokines and the RABV N gene. Graph-Prism® 5.0 and Instat® softwares were used as analysis tools. Gene expression of cytokines/chemokines were first compared between treated and non-treated groups inoculated with the same variant, at the same period, 5 or 10 days p.i (ex: hv3 non-treated 5 days vs hv3 siRNA treated at 5 days); a second comparison was made between groups at the same condition, inoculated (with same viral variant) treated and non-treated but at different periods, 5 and 10 days (ex: hv2 siRNA treated at 5 days vs hv2 siRNA treated at 10 days).
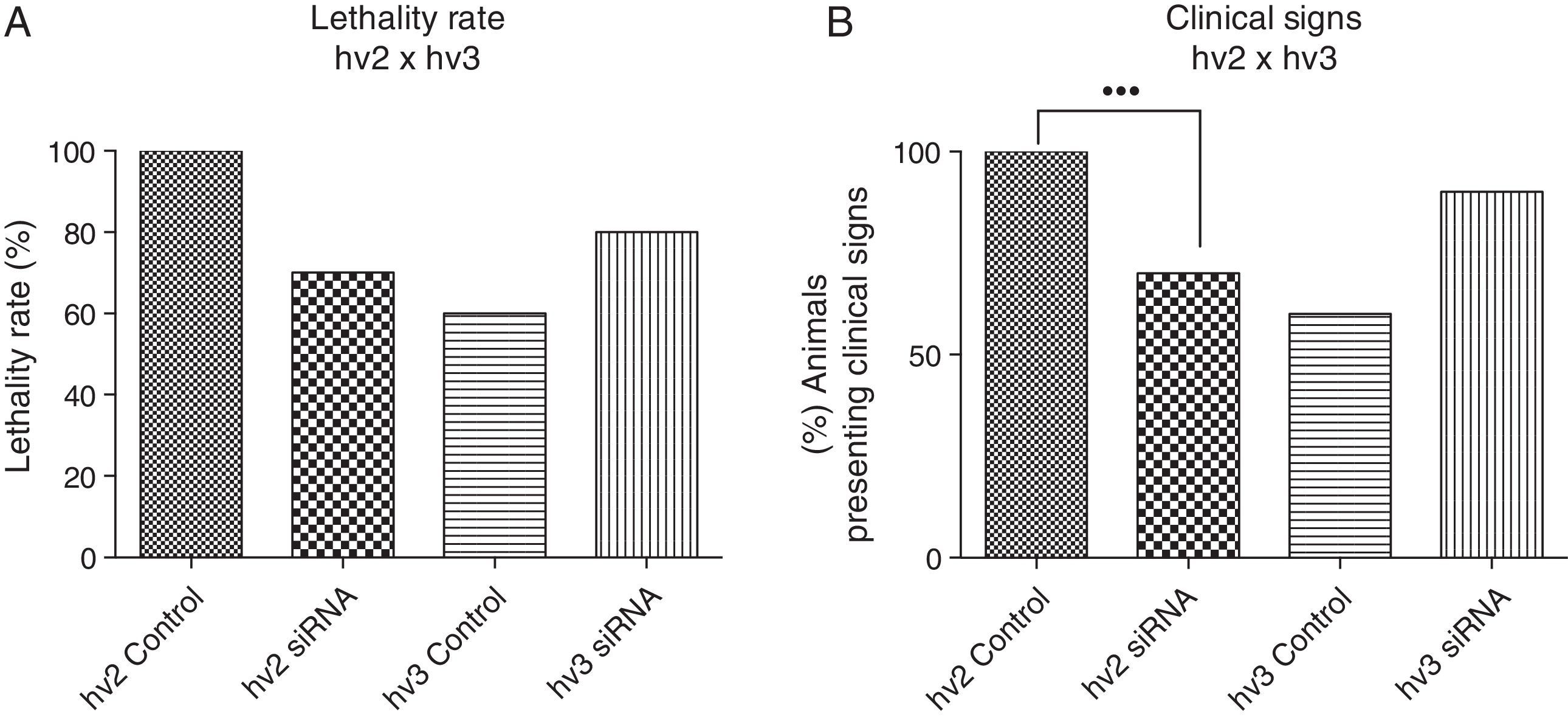
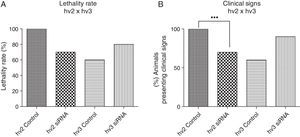
Results and discussionThere was a nonsignificant statistical difference in mortality rate in the groups treated with siRNAs irrespective of the variants used. For hv2, the lethality rate was 100% in non-treated and 70% in treated groups (p=0.27; HR=0.57); for hv3, the lethality was 60% in non-treated and 80% in treated groups (p=0.21; HR=1.97) (Fig. 1A).
(A) Lethality rate of controls and siRNA-treated groups inoculated with variant 2 (hv2) and variant 3 (hv3); Cox proportional hazards were used to estimate lethality rates and hazard ratios between groups. No statistical difference was found. (B) Percentage of animals in each group showing clinical signs, which included weight loss, ruffled fur, hunched back, hypoexcitability, hyperexcitability, paralysis, and tetraplegia; Kruskal–Wallis test showed a statistical difference (p=0.0006) between the hv2 control and siRNA-treated group.
Clinical evaluation of animals infected with variant 2 and treated with siRNA showed less severity of clinical disease, which included weight loss, paralysis, and death (p=0.0006) compared to the controls. However, no clinical difference was observed among hv3-infected animals either treated or non-treated with siRNA (Fig. 1B).
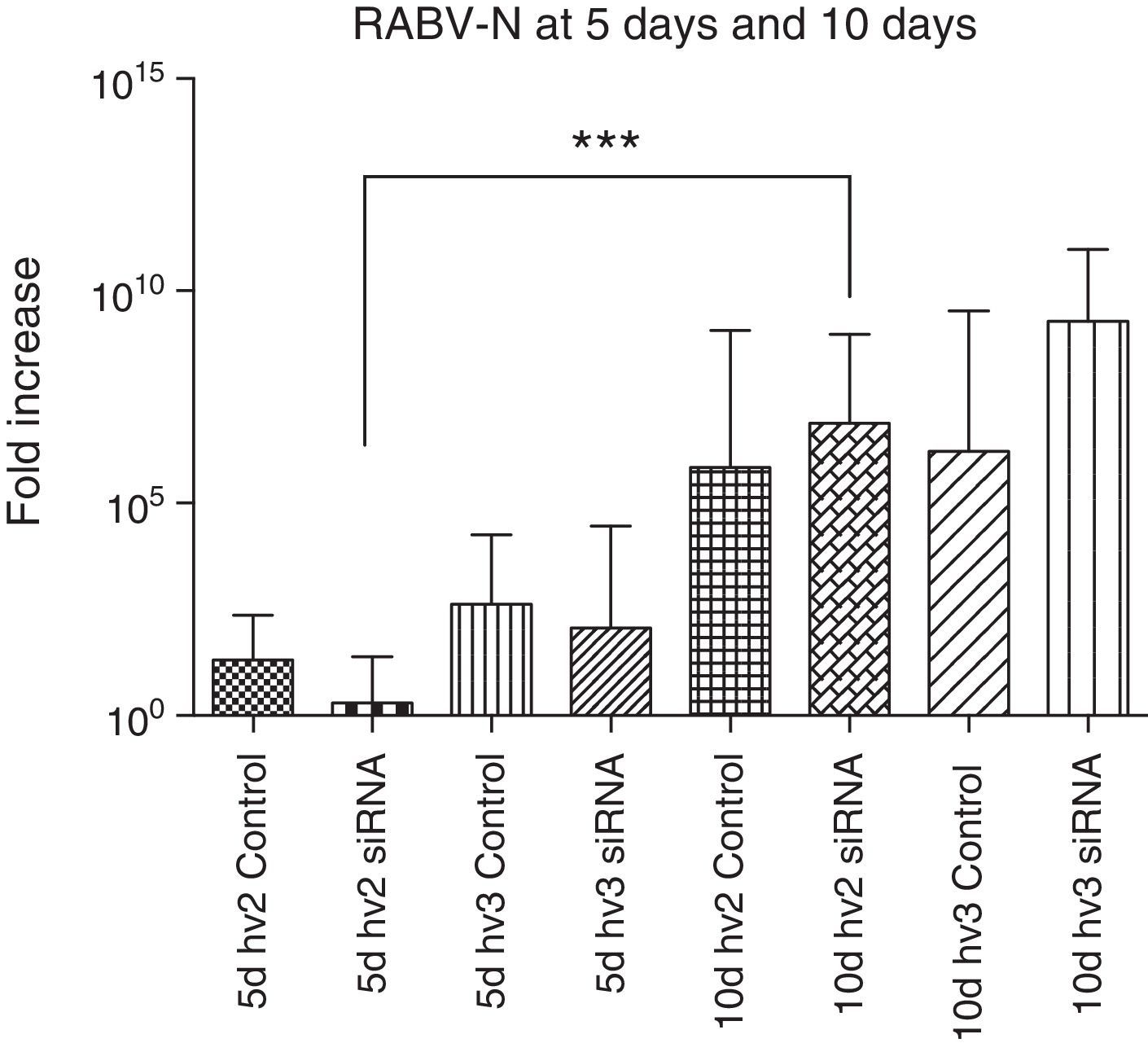
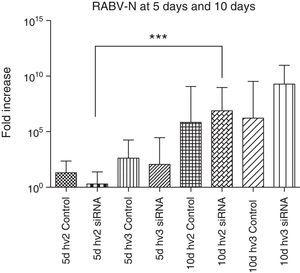
The N gene expression of all groups compared at the same period, either at 5 or 10 days p.i., showed no significant difference. However, N gene expression in the hv2 treated group was significantly increased at day 10 vs day 5 p.i. (p<0.0001). The increase in N gene expression is expected to follow disease progression.28 However, despite not reaching statistical significance, hv2 siRNA-treated animals at day 5 had very low expression compared to all other groups at the same period, reflecting some interference of siRNA in virus replication in this study (Fig. 2).
RABV N gene expression in the brain of mice infected with hv2 or hv3, treated or non-treated (control) with siRNA. Kruskal–Wallis test was applied to compare the results between different groups at day 5 and day 10 p.i. There was no difference between the hv2 and hv3 groups at day 5 or at day 10. However, hv2 siRNA-treated groups showed a significant difference (***p<0.001) at 5 vs 10 days.
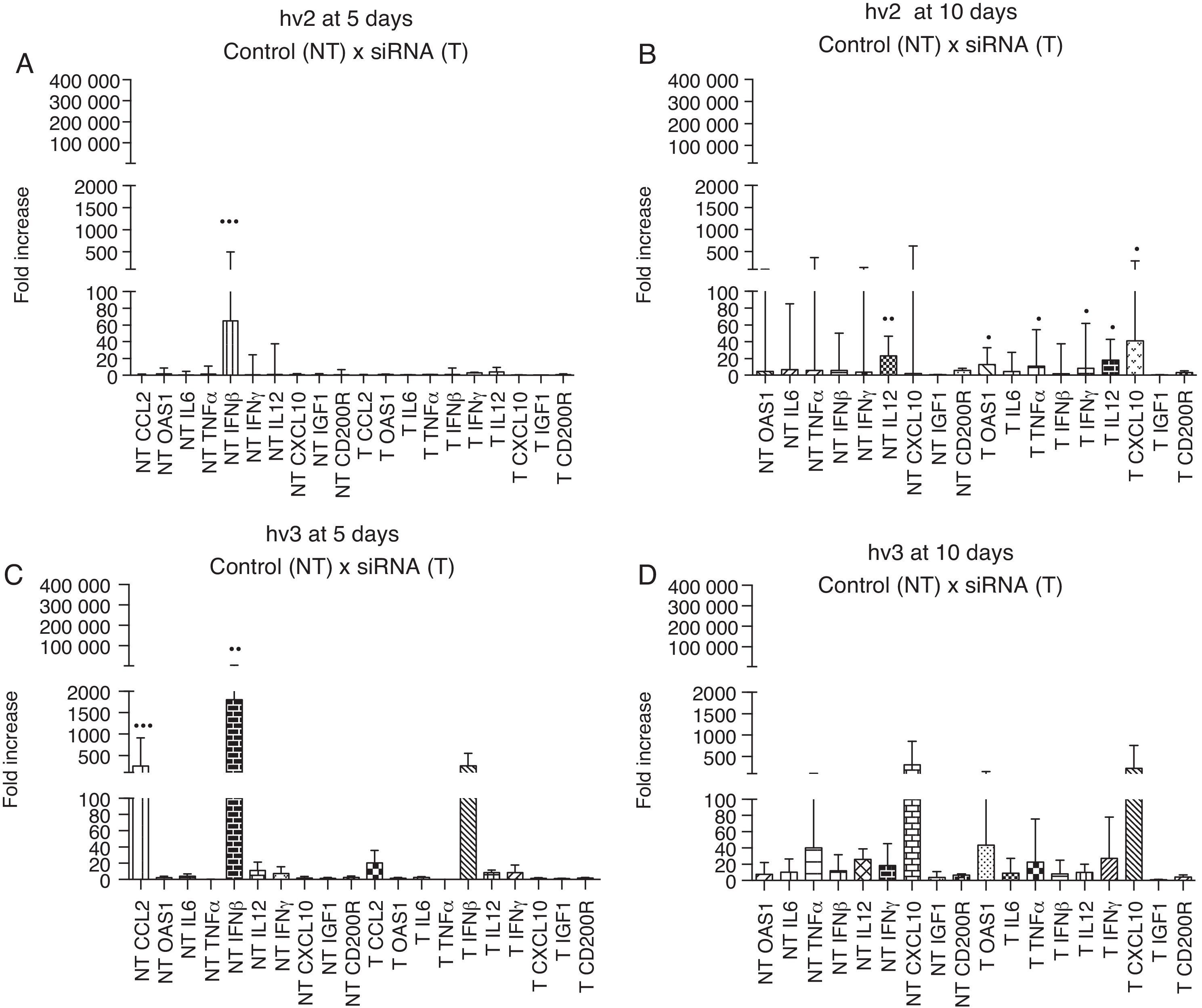
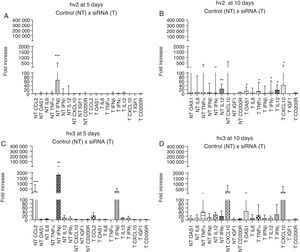
The brain immune response of different groups showed high expression of IFN-β (p<0.001) at day 5 p.i. in hv2 non-treated group and IFN-β (p<0.01) and CCL2 (p<0.001) in hv3 non-treated group; at day 10 p.i. hv2 non-treated group showed increased IL12 expression (p<0.01). There was no statistical difference in the analyzed immune markers in the treated hv2 group at day 5, whereas at day 10 OAS1, TNF-α, IL12, IFN-γ, and CXCL10 were increased (p<0.05) (Fig. 3). Infected cells showed a rapid production and release of IFN-β, which is important for host survival. This induces the expression of several IFN-stimulated genes (ISGs), such as OAS1, that exert an antiviral effect similar to IFN-β, but at different stages of viral replication.28 Damaged neurons also produce IFN-γ, IL1, IL6, IL12, CCL2, CCL4, CCL5, CCL7 and the IFN-inducible protein, CXCL10. All these cytokines and chemokines are responsible for the upregulated expression of major histocompatibility complex (MHC) molecules on the surface of microglia and also for the increased expression of adhesion molecules by endothelial cells. All the associated factors are important for induction of the adaptive immune response, which involves the activation and migration of T cells, as well as the production of specific antibodies.30–32 Enhanced N gene expression at day 10 vs day 5 in hv2-infected mice may indicate that at day 5 siRNA interfered with viral replication, which was associated with no difference among cytokine/chemokine gene expression at day 5 in this group (e.g., IFN-β), suggesting that viral levels were not sufficient to trigger the host immune response. This pattern of response did not occur in the hv2 non-treated group that showed a higher IFN-β expression at day 5 nor in the hv3 treated group, which has a similar gene expression profile compared to non-treated groups.
Relative gene expression of cytokines and chemokines in different groups (hv2 and hv3) at day 5 and day 10 p.i. The Kruskal–Wallis test was applied to analyze the results. (A) Expression at day 5 p.i. in the hv2 control (NT) and siRNA-treated (T) groups; IFNβ was highly expressed in the NT group (***p<0.001). (B) Expression at day 10 p.i. in the hv2 control (NT) and siRNA-treated (T) groups; IL12 was highly expressed in the NT group (**p<0.01); in the treated group, OAS1, TNFα, IL12, IFNγ and CXCL10 expression levels were increased (*p<0.05). (C) Expression at day 5 p.i in the hv3 control (NT) and siRNA-treated (T) groups; IFNβ was highly expressed in the NT group (**p<0.01). (D) Expression at day 10 p.i. in the hv3 control (NT) and siRNA-treated (T) groups; no significant difference was found.
Considering the differences in the results between variant 2 and 3, the identity of siRNA sequences and the bat and dog viruses used in this study were blasted showing identity ranging from 95 to 100% for hv2 and 86 to 95% for hv3. The siRNAs tested were designed based on Paster virus (PV),22 which is a fixed strain, and the identity results indeed showed a difference between street rabies virus and siRNA sequences, with lower identity for the bat variant.
Although the N gene is considered a conserved site, studies have shown a significant genetic variability in street rabies virus strains, from 5 up to 49%.33,34 It is important to remember that almost a perfect complementary sequence between siRNA molecule and the viral RNA target is necessary to induce cleavage of mRNA.35 This requirement may explain the difference found in the results for dog and bat variants observed in this study and also the lack of difference in the N gene expression among the groups evaluated in the same period.
To assess if treatment with siRNA could at least be partially effective in the symptomatic phase of the disease, an additional study in mice was developed administering a single dose of siRNAs only after the onset of clinical signs. No difference in mortality, clinical evaluation, or prolongation in the evolution period was observed in any of the treated groups (data not shown).
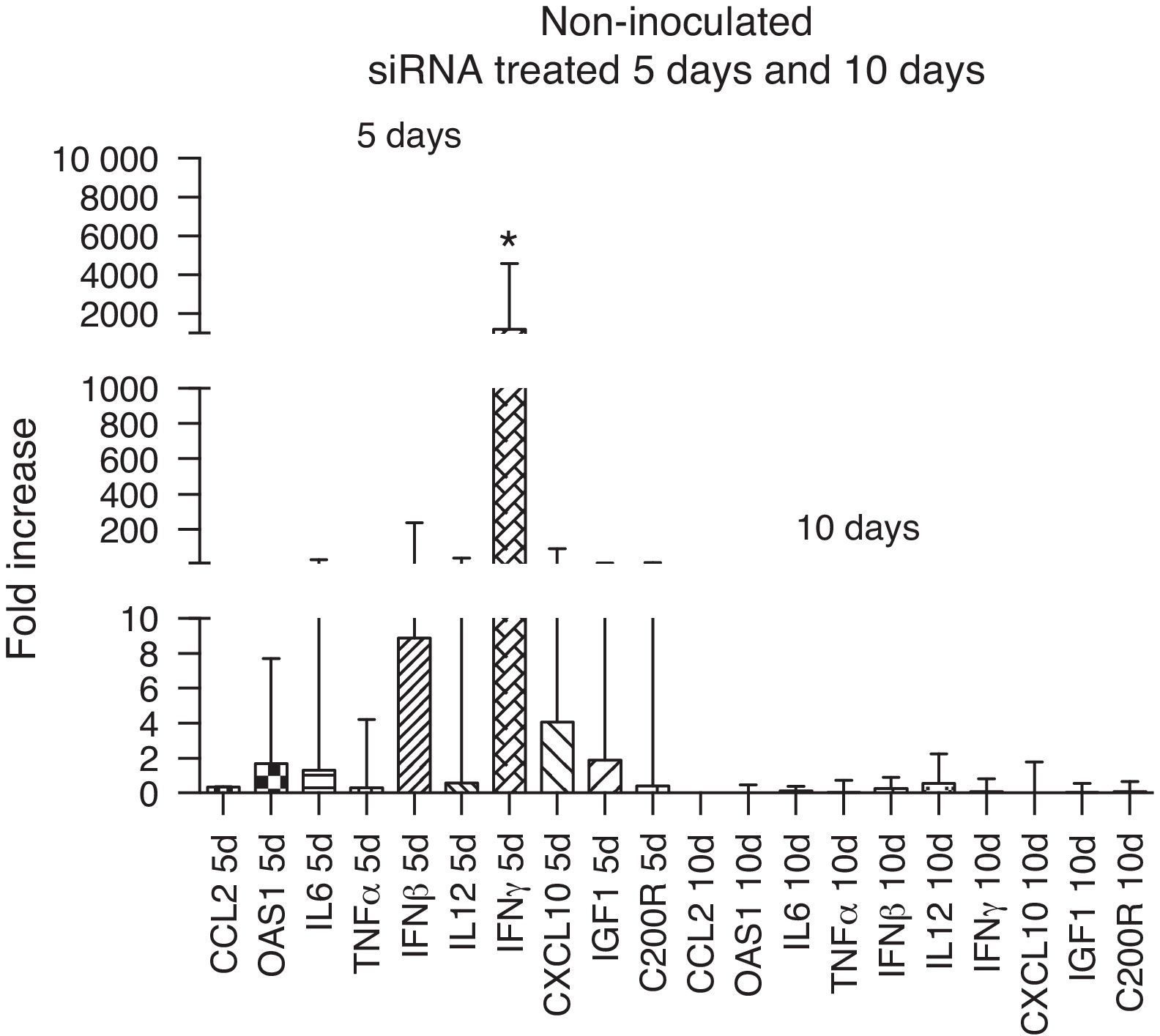
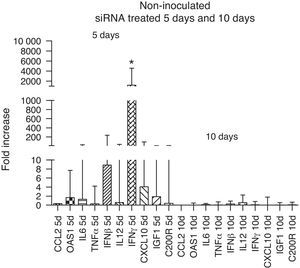
The immune response evaluation in non-inoculated animals treated with siRNA showed an up-regulation of all immune markers, in particular that of IFN-γ at day 5 compared to day 10. This increase disappeared at 10 days, reflecting the reduction in the activity of siRNA, which can last up to 6 days36 (Fig. 4).
ConclusionTherapy with siRNA has neither reduced the lethality rate in two different street rabies virus infections nor N gene expression. However, there were less severe clinical signs using siRNA therapy in variant 2 infection. The efficacy of siRNA therapy is closely associated with the identity of the siRNA design and its target. In this study, a higher identity was reached with variant 2 despite variant 3, justifying the obtained results. A reduced expression of immune markers with siRNA therapy in infected, but not in non-infected mice, may have occurred as a result of the antiviral effect. However, the results were not translated on reduced mortality or expression of N gene, which can indirectly reflect virus replication suggesting that pre-designed siRNA may not be useful in rabies treatment. The potential in applying this technology is limited because in medical practice the type of virus infecting the patient is unknown in almost 100% of the cases. More studies are necessary to overcome this limitation and to show whether this technology could be applied either alone or associated with other therapeutic measures.
FundingFundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP – No. 08/11446-1) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 482726/2012-1).
Conflicts of interestThe authors have declared there is no conflict of interest.
We thank Dr. Maria Luiza Carrieri and Dr. Ivanete Kotait for kindly providing both rabies virus strains.