Rahnella aquatilis is an environmental Gram-negative bacillus that is rarely reported as human pathogen, being mainly associated with infections in immunocompromised patients. Herein we describe two cases of R. aquatilis isolates recovered from endotracheal aspirate cultures of different patients in a tertiary hospital located in the city of São Paulo, Brazil. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and 16S rDNA gene sequencing were performed to confirm bacterial identification after the isolates being erroneously identified as Pantoea spp. by automated system. Both isolates showed the same PFGE pattern and presented the β-lactamase encoding gene blaRAHN-1, responsible for resistance to cephalothin. The isolates were susceptible to broad-spectrum cephalosporins, carbapenems, fluoroquinolones, aminoglycosides, and polymyxin B. This report shows the presence and transmission of uncommon bacteria in the nosocomial environment and alerts us about the need for new tools of correct microbiologic diagnosis.
Rahnella spp. is a facultative anaerobic and nitrogen fixer member of the Enterobacteriaceae family.1 This genus was described in 1976, and was initially denominated as a Group H of Enterobacteriaceae, based on numerical taxonomy. With the advent of molecular techniques, this new genus was renamed Rahnella. It has been mainly isolated from water and soil, and therefore it can be abundantly found in fresh products. The Rahnella genus comprises R. aquatilis, R. genomospecies 2 and R. genomospecies 3, that can be differentiated only by molecular techniques.2 To date, few cases of infection caused by R. aquatilis have been reported in literature, affecting mainly immunocompromised patients.1,3 In 2001, Bellais and colleagues reported a novel non-inducible and chromosomally encoded Ambler class A β-lactamase, named RAHN-1, which is intrinsic of R. aquatilis strains.4 Two variants of blaRAHN have been described to date (blaRAHN-1 and blaRAHN-2), and contribute to one of the few resistance mechanisms described in this pathogen. Although clinical and environmental RAHN-producing R. aquatilis strains reported thus far were usually resistant to penicillins and narrow-spectrum cephalosporins, these strains remained susceptible to ceftazidime, imipenem, and were inhibited by clavulanic acid.5
The identification of R. aquatilis by classical microbiologic features is difficult, since there is not a single biochemical test that could differentiate R. aquatilis isolates from other Enterobacteriaceae.6R. aquatilis is a non-motile bacillus at 35°C (motile only at 25°C), use citrate as the carbon source, and is able to ferment main carbohydrates, after 48h incubation at 35°C. However, in MacConkey agar, a few weak lactose colonies are usually observed.2R. aquatilis maybe misidentified as Enterobacter agglomerans, since these bacteria species share similar biochemical profile. Thus, correct identification by molecular techniques maybe required. In the last years, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has been established as a preferable alternative method for accurate bacterial identification. Herein, we report the microbiological features of RAHN-1-producing R. aquatilis isolates recovered from two patients admitted to a tertiary hospital in the city of São Paulo, Brazil.
Two different Gram-negative rods were recovered from endotracheal aspirate cultures of distinct hospitalized patients, in the same period, at a neurological intensive care unit (ICU). In the first case, a 64 y-o male hospitalized for a long-term due to autoimmune encephalopathy associated to Lyme disease was submitted to tracheostomy due to respiratory disorders. During his stay, he developed various infections (respiratory and urinary) and received several courses of antimicrobial therapy. However, the patient had not been on antimicrobial therapy in the month before R. aquatilis isolation. After being hospitalized for five months, the patient was diagnosed with a pulmonary infection due to the presence of purulent endotracheal aspirate and radiological image compatible with parenchymal lung infiltrate. Pseudomonas aeruginosa and Pantoea spp. were identified by Vitek®2 automated system (bioMérieux, Marcy l’Etoile, France) in the endotracheal aspirate specimen, both at >105UFC/mL. Meropenem therapy was introduced and clinical improvement was observed seven days thereafter. In the second case, a 37 y-o male with a degenerative neurological syndrome (Hallervorden-Spatz syndrome) admitted to a long-term institution developed septic shock due to pulmonary infection in the left lower lobe, requiring mechanical ventilation and vasoactive drug support. Antimicrobial therapy with teicoplanin and meropenem was initiated with clinical improvement after one week. No pathogen was recovered from any clinical samples collected after improvement. However, after two-month several bacterial isolates were cultured from blood samples in a period of one month, including a KPC-producing Klebsiella pneumoniae, following by KPC-producing Enterobacter aerogenes and Staphylococcus haemolyticus. The patient was treated with multiple courses of antimicrobial agents including tigecycline, amikacin, teicoplanin, and meropenem. One week after finishing antimicrobial therapy the patient presented with fever and a carbapenem-resistant P. aeruginosa and Pantoea spp. isolates were recovered from endotracheal aspirate. The patient received a new course of tigecycline and amikacin therapy with clinical improvement.
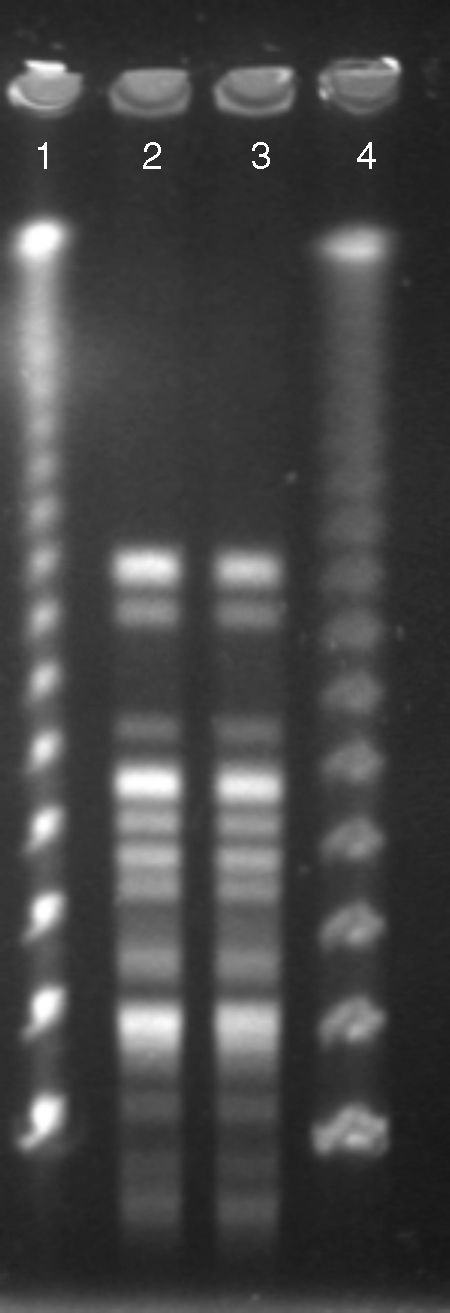
Both Pantoea spp. isolates were referred to a reference laboratory for confirmation of bacterial identification and antimicrobial susceptibility. Mass spectrometry by Vitek® MS (bioMérieux S/A, Marcy I’Etoile, France) identified both isolates as R. aquatilis (score of 99%) and 16S rDNA gene sequencing confirmed those results with 100% similarity and coverage. Rahnella species were recently included in the commercial mass spectrometry databases, thus, allowing their identification from clinical and environment samples.7 To determine the clonal relationship, pulsed-field gel electrophoresis (PFGE) using SpeI as the restriction enzyme was performed as previously described.8 The analysis was performed according to the Tenover criteria.9 The Rahnella isolates belonged to a single PFGE patterns (Fig. 1), suggesting that a patient-to-patient transmission had occurred, since they were admitted to the same ICU and shared the same hospital health care staff in the same period of time. Caroff and colleagues described one of the first transmission of R. aquatilis in nosocomial environment.10 In that study, because the patients were unrelated it was suspected that prolonged parental nutrition given to by both patients was the source of spread. Other studies identified the water used in the bronchial washing process and citrate solution used in parenteral nutrition solution as the main sources of contamination by R. aquatilis.3,6,10 Fortunately, no further transmission of R. aquatilis has been observed and investigation of the source was not evaluated.
Antimicrobial susceptibility was evaluated by determining the minimal inhibitory concentrations (MICs) using broth microdilution according to the CLSI guideline.11 The antimicrobial agents tested were ampicillin, amikacin, gentamicin, cefepime, cephalothin, ceftriaxone, ceftazidime, aztreonam, meropenem, imipenem, ciprofloxacin, levofloxacin, and polymyxin B. According to CLSI breakpoints,12 both isolates were susceptible to levofloxacin (MIC, ≤0.25μg/mL), ciprofloxacin (MIC, ≤0.125μg/mL), gentamicin (MIC, ≤0.125μg/mL), amikacin (MIC, ≤0.5μg/mL), ampicillin (MIC, 8μg/mL), cefepime (MIC, 0.25μg/mL), ceftriaxone (MIC, 1μg/mL), aztreonam (MIC, 1μg/mL), imipenem (MIC, 0.25μg/mL), meropenem (MIC, ≤0.06μg/mL), and polymyxin B (MIC, 4μg/mL). Since the isolates were resistant only to cephalothin (MIC, 128μg/mL), the presence of β-lactamases encoding genes was investigated by PCR followed by DNA sequencing using the ABI3500 Genetic Analyzers (Applied Biosystems, Foster City, CA). The presence of blaRAHN-1 gene was detected in both isolates, justifying resistance to cephalothin observed in these isolates. Although the two R. aquatilis isolates were susceptible to broad-spectrum cephalosporins, previous studies have demonstrated that RAHN-1 shows hydrolytic activity against cefotaxime and ceftriaxone.4 So far, all isolates of Rahnella spp. reported in the literature, were susceptible to most antimicrobials agents tested. Thus, broad-spectrum cephalosporins are usually employed for the treatment of infections caused by these pathogens. In contrast, in some cases when R. aquatilis is isolated with another pathogen, the antimicrobial treatment must take into account the coverage against both pathogens.
Human infections caused by R. aquatilis are infrequent and had not been previously reported in Brazil, which could be justified, at least in part, by the inability of automated systems to identify environmental microorganisms. Since R. aquatilis biochemical profile is very similar to that of other Enterobacteriaceae, like Pantoea spp. and Enterobacter spp., more reliable techniques are crucial to identify environmental isolates from clinical specimens in order to better define the epidemiological role played by these species in the nosocomial environment. In this context, MALDI-TOF MS seems to be a reliable tool in the identification of such isolates. Due to the isolation of multi-drug resistant P. aeruginosa strains in the same clinical specimens where R. aquatilis were also isolated, it is difficult to define the exact role of R. aquatilis as the pathogen responsible for ventilator-associated pneumonia. However, the isolation of environmental multisusceptible bacteria in the hospital setting could represent a real threat, since, generally, this microorganisms carried virulent determinants and are able to easily acquire resistant genes carried by motile genetic elements from other common pathogens. Thus, the emergence of such microorganisms as clinically significant pathogens, especially in immunocompromised patients, is worrisome.
Conflicts of interestA.C.G. recently received research funding and/or consultation fees from AstraZeneca, MSD, Novartis, Thermo Fisher Scientific, and BioMérieux. Other authors have nothing to declare.
We are grateful to Fernanda Rodrigues-Costa and Ioná Torres Magalhães for their excellent technical contribution to the manuscript.