Dengue has been a significant public health problem in Colombia since the simultaneous circulation of the four dengue virus serotypes. The replicative fitness of dengue is a biological feature important for virus evolution and contributes to elucidating the behavior of virus populations and viral pathogenesis. However, it has not yet been studied in Colombian isolates. This study aimed to compare the replicative fitness of the four dengue virus serotypes and understand the association between the serotypes, their in vitro infection ability, and their replication in target cells. We used three isolates of each DENV serotype to infect Huh-7 cells at an MOI of 0.5. The percentage of infected cells was evaluated by flow cytometry, cell viability was evaluated by MTT assay, and the pathogenicity index was calculated as a ratio of both parameters. The replicative fitness was measured by the number of viral genome copies produced using quantitative PCR and the production of infectious viral progeny was measured by plaque assay. We showed that Huh-7 cells were susceptible to infection with all the different strain isolates. Nevertheless, the biological characteristics, such as infectious ability and cell viability, were strain-dependent. We also found different degrees of pathogenicity between strains of the four serotypes, representative of the heterogeneity displayed in the circulating population. When we analyzed the replicative fitness using the mean values obtained from RT-qPCR and plaque assay for the different strains, we found serotype-dependent behavior. The highest mean values of replicative fitness were obtained for DENV-1 (log 4.9 PFU/ml) and DENV-4 (log 5.28 PFU/ml), followed by DENV-2 (log 3.9 PFU/ml) and DENV-3 (log 4.31 PFU/ml). The internal heterogeneity of the replicative fitness within each serotype could explain the simultaneous circulation of the four DENV serotypes in Colombia.
Dengue is a public health problem in tropical and subtropical regions, where an estimated four billion people are at risk of suffering from the disease. In 2010, 390 million cases of dengue were estimated worldwide, of which 96 million presented with outward manifestations.1 Colombia is a hyperendemic country for dengue, and although the prevalence of serotypes has changed over time, the four DENV serotypes have circulated simultaneously in 30 of the 32 departments of the country. For more than 30 years, DENV serotype circulation has occurred without displacement of any serotype and with high variability between the departments of the country.2 The number of cases of dengue in Colombia is still high; during 2018, 44,825 cases were reported, of which 47.4% corresponded to dengue without warning signs, 51.4% to dengue with warning signs, and 1.2% to severe dengue.3 The course of the disease and the degree of clinical complications are related to the genetics and the immune response of the patient and the intrinsic characteristics of the infecting viral strain,4 which could explain the different epidemiological dynamics observed worldwide. Different factors contribute to the high incidence of the disease in the country, such as high population densities in rural areas with poor infrastructure, which favors the presence of the vector Aedes aegypti, fluctuation between seasons of drought and high rainfall, insufficient methods for vector control, poverty with the inherent difficulty of access to healthcare, an underdeveloped national system of diagnosis and clinical management, hyperendemicity and natural changes in the relative abundance of circulating DENV serotypes.5
Dengue virus (DENV) has a complex classification and belongs to the Flaviviridae family. This virus has four types (DENV-1, DENV-2, DENV-3, DENV-4) with 62–67% sequence identity.6 The classification in four serotypes is based on the immune response of the patient to primary infection by one of the serotypes. Primary infection protects against a secondary infection by a homologous DENV serotype but confers partial and transient protection against a heterologous DENV serotype.7 Additionally, each type of DENV is further classified into monophyletic taxonomic groups or genotypes, as well as subclades or lineages.8,9 To date, five genotypes of DENV-1, six of DENV-2, five of DENV-3, and five of DENV-410–14 have been described, and the relationship between their population dynamics and epidemiological behavior is still under investigation.
The epidemiological association between infectious DENV serotypes and the evolution of severe clinical symptoms is not conclusive. Previous studies suggest a correlation between geographic heterogeneity and spatial dependence of the circulating viral strain and the clinical outcome of DENV disease. A meta-analysis found that the most pathogenic serotypes in southern Asia are DENV-3 followed by DENV-4, DENV-2, and DENV-1, while in countries including Peru, Cuba, France, Colombia, and Brazil, DENV-2 was described as the most pathogenic strain followed by DENV-3 and DENV-4.15 A study in Nicaragua described DENV-3 as the most pathogenic and DENV-4 as the least pathogenic serotype.16
The replicative fitness of a virus population demonstrates its ability to produce infectious progeny in a specific microenvironment.17 Changes in replicative fitness profiles may favor or impair the transmission of the virus. The difficulty of evaluating the replicative fitness of a viral population during the natural transmission cycle has led to the development of in vitro systems that quantify viral titers as an approximation to replicative fitness. This approach has been used in different studies to understand the epidemiological dynamics of DENV in viral population replacement events. In Nicaragua, a clade replacement of the DENV-2 Asian-American genotype clade NI-1 by clade NI-2B was reported in 2009. This replacement was related to a higher replicative fitness of strains belonging to the NI-2B clade, which was evaluated in C6/36 mosquito cells and iDCm human-derived monocytes.18 In Puerto Rico in 2011, there was a displacement of clade PR-1 by clade PR-2B belonging to DENV-2. This event was associated with a differential fitness between the clades when the replicative fitness was evaluated in human liver cells (Huh-7). Additionally, clade PR-2B had higher fitness as demonstrated by viral inhibition of the interferon type I response.19
In countries where the four DENV serotypes are in cocirculation, such as Colombia, few studies have been carried out to understand the population dynamics and their possible epidemiological relationship. In Colombia, a study evaluated the replicative fitness of DENV in C6/36 mosquito cells and colonies of A. aegypti using a viral strain of each DENV serotype and described a differential fitness between serotypes, in which DENV-2 had the highest replicative capacity followed by DENV-1, DENV-4, and DENV-3.20 However, the use of one strain per DENV serotype might not be representative of the fitness of other viral circulating subpopulations. Additionally, different evolutionary patterns have been described between viral populations that replicate in mammalian and mosquito cells. Viral progenies produced from mammalian cells such as Huh-7 cells accumulate a higher number of mutations compared to C6/36 cells.21 These mutations lead to an increase in the genotypic diversity of a viral population, and this diversity has a direct relationship with variations in fitness that would facilitate the transmission cycle.22,23
In this study, we evaluated the biological behavior and compared the replicative fitness of Colombian strains of the four DENV serotypes using three clinical isolates from each serotype and the human hepatocyte cell line Huh-7 as a model of infection. We found that viral characteristics such as percentage of infection, effects on cell viability, and the pathogenicity index are dependent on the strain and not on the serotype, while the replicative fitness is influenced by the serotype and has an internal heterogeneity between strains that is characteristic of a natural dengue population. From the analysis of our results, we were able to define one group, DENV-1 and DENV-4 strains, with higher replicative fitness, and a second group, DENV-2 and DENV-3 strains, with lower replicative fitness. This study constitutes the first report of a complete evaluation of the replicative fitness of DENV strains of each serotype in Colombia, a hyperendemic country where the control and management of the disease are complicated by the simultaneous circulation of the four DENV serotypes.
Materials and methodsCell culture and virusA. albopictus C6/36HT (HT- high temperature) cells, immortalized human liver (Huh-7) cells and baby hamster kidney fibroblast (BHK-21) cells were maintained in Dulbecco´s Modified Eagle´s Medium (DMEM, Sigma-Aldrich, St. Louis, MO, USA) and supplemented with 10% fetal bovine serum (Fetal Bovine Serum (FBS), Gibco, Sigma-Aldrich, St. Louis, MO, USA). Huh-7 and BHK-21 cells were grown at 37°C and 5% CO2. C6/36HT cells were grown at 33°C and 5% CO2. The virus strains of DENV-1 (DENV-1 116, DENV-1 S33, DENV-1 S24), DENV-2 (DENV-2 216, DENV-2 S3, DENV-2 209), DENV-3 (DENV-3 S7, DENV-3 S8, DENV-3 S26) and DENV-4 (DENV-4 416, DENV-4 S29, DENV-4 S32) were isolated from patients or healthy donors and propagated in C6/36HT cells as previously described.24 The low-passage virus stocks were inoculated in C6/36HT, harvested 3 days post-infection, aliquoted and stored at —80°C until use. For this study, previously standardized viral isolates after 10 passages (DENV-1 OMS, DENV-2 INS16, DENV-3 OMS, DENV-4 INS strains) were included as positive controls for plaque assay.
Cell infectionTwenty hours prior to infection, Huh-7 cells were seeded in 48-well plates at a density of 2.5×104 cells per well. The cells were infected at MOI 0.5 and incubated for 1h at 37°C in a 5% CO2 atmosphere. Then, the inoculum was removed, and DMEM supplemented with 10% fetal bovine serum (FBS, Gibco) was added. The cells were further incubated for 72h at 37°C in 5% CO2 and the supernatants were collected and stored at −80°C until use.
Detection of viral antigen in Huh-7 cells infected with DENVThe susceptibility of Huh-7 cells to infection by DENV was evaluated by an immunoperoxidase assay by detection of viral protein E. Briefly, 20h before infection, Huh-7 cells were seeded on SPL Life Sciences round coverslips at a density of 2.5×104 per well and infected for 48h at an MOI 1 with the viral strains DENV-1 116, DENV-2 216, DENV-3 S8 and DENV-4 416. Cells were fixed with 4% paraformaldehyde for 30min and then permeabilized with 0.3% Triton X-100 at room temperature. Inactivation of endogenous peroxidase was performed with 0.25% H2O2 in 50% methanol, and nonspecific binding sites were blocked with 10% goat serum. The cells were incubated with the DENV monoclonal 4G2 antibody at a 1:800 dilution for one hour at 37°C, washed and incubated with a peroxidase-coupled anti-mouse IgG. Finally, the reaction was developed with 0.02% H2O2 and 0.075% diaminobenzidine in 0.1M Tris-HCl buffer at pH 7.2 and photographed for cell counting under an inverted Axiovert 40 microscope.
Evaluation of the percentage of infected cells by flow cytometryTwenty hours prior to infection, 1×104 Huh-7 cells per well were seeded in 96-well microplates and maintained in DMEM supplemented with 2% FBS and 0.5g/ml gentamycin. Huh-7 cell monolayers were infected at MOI 0.5, as mentioned above for 72h, and processed for cytometry. Briefly, cells were treated with BD Cytofix/Cytoperm™ Plus Fixation/Permeabilization Solution Kit with BD GolgiPlug™ according to the manufacturer´s instructions and then washed and incubated with 4G2-FITC antibody (donated by the Functional Genetics of Infectious Diseases Unit, Institute Pasteur, Paris) for 30min at 4°C. After incubation, cells were washed once with PBS (pH 7.2), resuspended, and then analyzed using a FACScan flow cytometer (BD Accuri™ C6). Mock-infected cells were used as negative controls. At least 1000 cells were gated by light scatter. Data analysis was performed with FlowJo 10 software.
Cell viabilityCell viability was assessed using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) method. Seventy-two hours after infection, Huh-7 cell monolayers were incubated with 100μL MTT (0.5mg/ml) for 1h at 37°C in a 5% CO2 atmosphere protected from light. After incubation, the formazan salts were dissolved with dimethyl sulfoxide for 10min. The absorbance measurements were made in a TECAN Infinite M200pro reader at 450nm. Uninfected cells were used as a negative control, and cells treated with 0.1% saponin for five minutes were used as a positive control.
Estimation of the pathogenicity indexThe pathogenicity index was calculated as the ratio between the cell viability values and the previously reported percentage of infection for each strain. Strains with ratios >1 were classified as cytopathic, and strains with <1 value were classified as pathogenic.
Replicative fitness in Huh-7 cellsTwenty hours prior to infection, 2.5×104 Huh-7 cells per well were plated in a 48-well plate in medium supplemented with 2% FBS and gentamycin. Then, the cells were infected with each virus strain at MOI 0.5 in triplicate in two independent experiments. Supernatants were removed 72h after infection and stored at −80°C for subsequent quantification of viral genomic copies by RT-qPCR and viral infectious particles by plaque formation assay. The monolayers were used to determine cell viability by MTT.
Quantification of viral RNAViral RNA in the supernatants was isolated using a Stratec molecular kit (San Francisco, CA, USA). Viral RNA was reverse-transcribed for 60min at 37°C using M-MLV reverse transcriptase (Promega, USA) and random primers. qPCR was performed using the DyNAmo HS SYBR Green qPCR kit (Thermo, Massachusetts, USA) according to the manufacturer´s instructions using previously described primers.25 The qPCR conditions were as follows: 95°C for 15min, followed by 40 cycles of 95°C for 15s, 58°C for 1min and 72°C for 1s, followed by a melting curve analysis. For absolute quantification, a standard curve was obtained from ten serial dilutions of each titrated serotype. The viral titer for each strain was previously determined by plaque assay. RNA from each dilution was isolated, and standard curves with a minimum of four dilutions were set up.
Plaque assayBHK-21 cells were seeded at a density of 1×105 cells/well for the titration of serotypes 2 and 4 and 8×104 cells for serotypes 1 and 3 per well in a 24-well plate. The cell suspension was infected with serial dilutions of each virus. Four hours after the infection, DMEM 2X containing 3% carboxy-methyl-cellulose (Sigma C4888-500G), 7.5% sodium bicarbonate (NaHCO3) and supplemented with 6% FBS and 0.5 μg/ml gentamycin was added. NaHCO3 was used to reach the optimal pH for each serotype: DENV-1 pH 8.0, DENV-2 pH 7.0, DENV-3 pH 7.6, and DENV-4 pH 7.4. The infected cells were incubated for seven days with DENV-1 and DENV-3, six days with DENV-4 and five days with DENV-2 at 37°C and 5% CO2. Then, the cells were fixed and stained with 1% crystal violet solution in 70% methanol for 25min. The crystal violet solution was washed, and the virus titer was calculated using the formula PFU/ml=(average number of plaques×dilution factor) / inoculum volume (μL).
DENV genotypingFor the determination of the genotype of each DENV strain, a region of the E gene of approximately 775 bp was amplified by PCR using the following program: 95°C for 3min, 30 cycles of 95°C for 30s, 53°C for 30s, 72°C for 30s and final extension for 5min at 72°C using previously reported primers.26 The PCR products were sequenced by Macrogen Inc. The results of the sequencing were analyzed using the database “DGV: Dengue Genographic Viewer”27
StatisticsThe data are presented as the median and interquartile range or mean±SD depending on the data. The data were analyzed by a Kruskal-Wallis test followed by a Dunn`s test of multiple comparisons or an ANOVA test and Tukey’s multiple mean comparisons test according to the normality of the data. All the data were analyzed using R 3.3.0 statistical software, and the graphics were made using Prism7.0a. Statistical significance was defined as p<0.05.
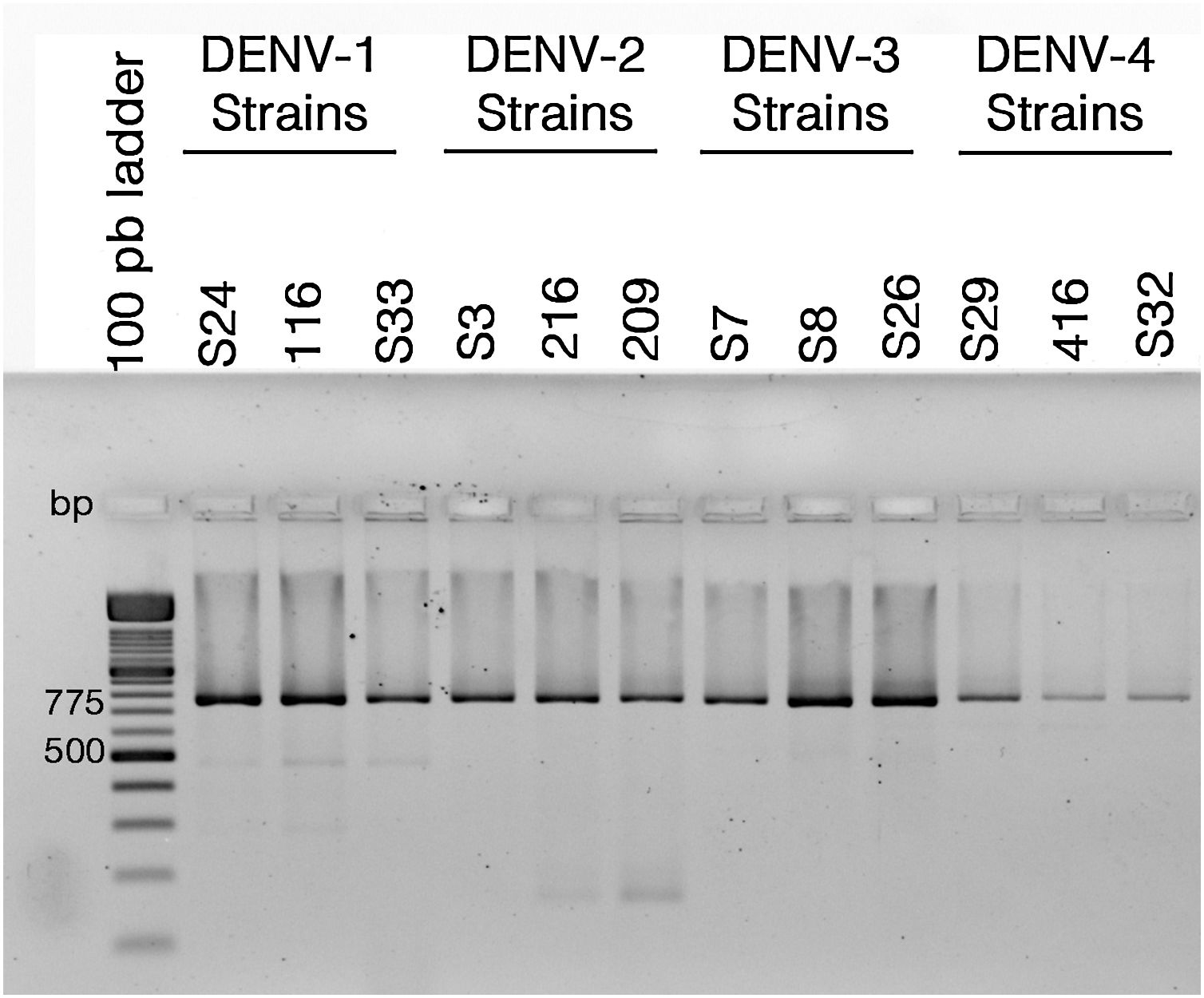
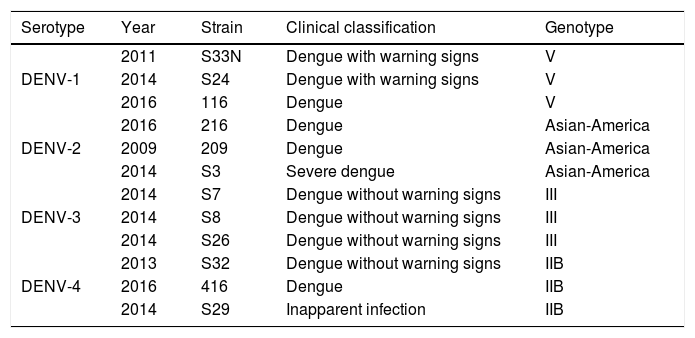
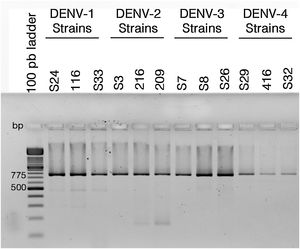
ResultsColombian DENV strains belong to the genotypes previously reportedThe 775 bp RT-PCR products were efficiently amplified from viral RNA from each of the strains used in this study (Fig. 1). Sequence analysis determined that all three DENV-1strains were classified in genotype V, the DENV-2 strains in the Asian-American genotype, the DENV-3 strains in genotype III and the three DENV-4 strains in genotype IIB (Table 1). These data agree with the molecular characterization of DENV previously reported for Colombia28–31 and demonstrate that the strains studied for each serotype belong to the same genotype, suggesting that the subsequent biological properties and replicative fitness analyses are influenced by strain and serotype characteristics and not by variations between genotypes as previously reported.32
Reverse transcriptase polymerase chain reaction (RT-PCR) with viral RNA from dengue virus clinical isolates for genotyping fragments of DENV-1 S24, DENV-1 116, DENV-S33, DENV-2 S3, DENV-2 216, DENV-2 209, DENV-3 S7, DENV-3 S8, DENV-3 S26, DENV-4 S29, DENV-4 416, DENV-S32. The amplification products of the expected size (775bp) were resolved in 2% agarosa gel stained with ethidium bromide.
Details of dengue virus strains.
| Serotype | Year | Strain | Clinical classification | Genotype |
|---|---|---|---|---|
| DENV-1 | 2011 | S33N | Dengue with warning signs | V |
| 2014 | S24 | Dengue with warning signs | V | |
| 2016 | 116 | Dengue | V | |
| DENV-2 | 2016 | 216 | Dengue | Asian-America |
| 2009 | 209 | Dengue | Asian-America | |
| 2014 | S3 | Severe dengue | Asian-America | |
| DENV-3 | 2014 | S7 | Dengue without warning signs | III |
| 2014 | S8 | Dengue without warning signs | III | |
| 2014 | S26 | Dengue without warning signs | III | |
| DENV-4 | 2013 | S32 | Dengue without warning signs | IIB |
| 2016 | 416 | Dengue | IIB | |
| 2014 | S29 | Inapparent infection | IIB |
The patients were located at Cundinamarca department, in the central region of Colombia, except for the strain S32 isolated from the Pacific region of Colombia.
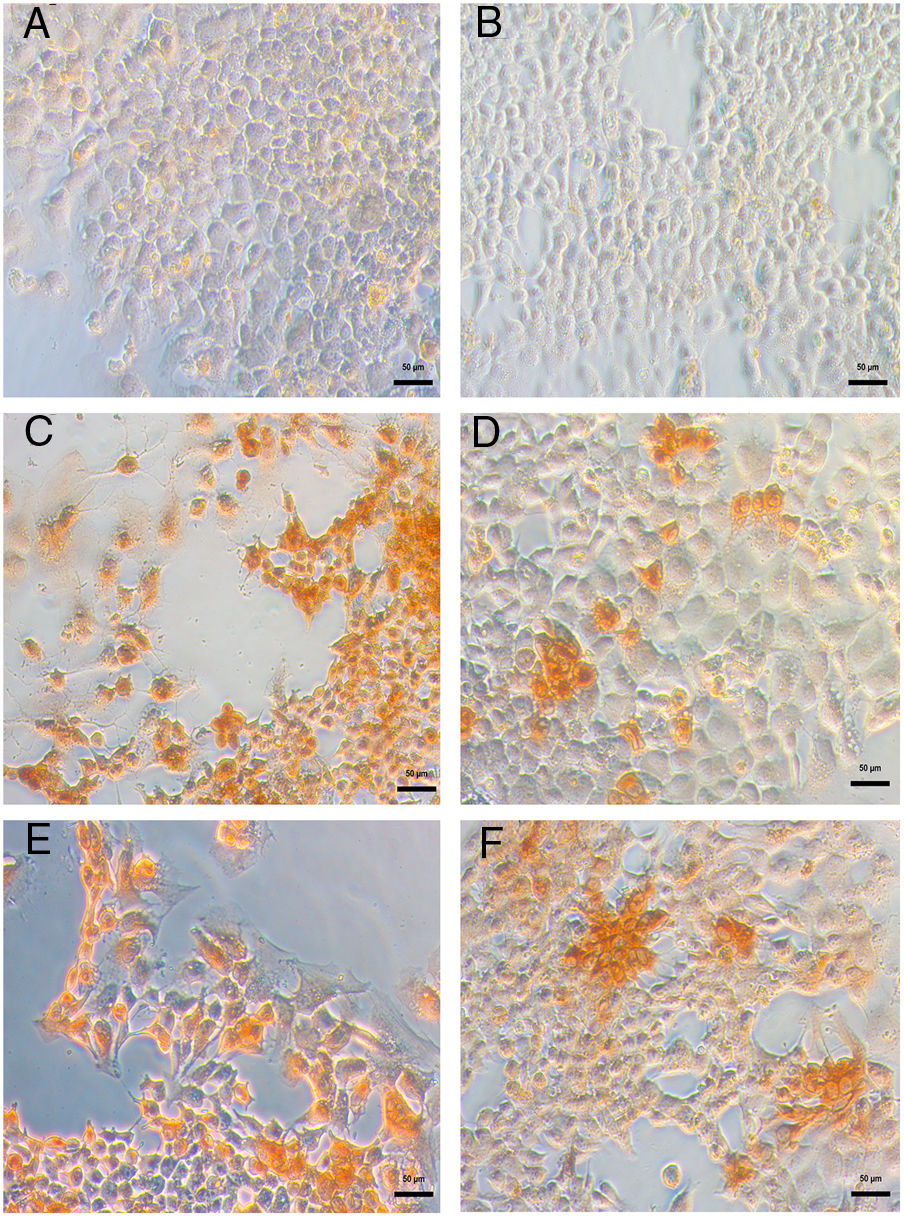
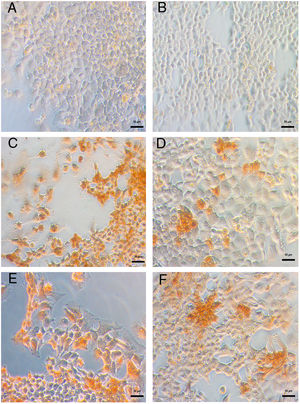
DENV infection in Huh-7 cells was confirmed by the detection of the viral E antigen in an immunocytochemistry assay. In cells infected with the four DENV serotypes, the presence of the viral E protein with a cytoplasmic distribution and no syncytia formation confirmed that Huh-7 cells are susceptible to DENV infection and represent a cell culture system that supports viral replication (Fig. 2). The production of infectious viral progeny after inoculation demonstrated that the infection had spread through the cell monolayer. The results showed that in addition to the importance of hepatocytes as a target of DENV infection in the pathophysiology of the disease, these cells are an adequate model for the study of the replicative fitness of DENV.
Huh-7 cells are permissive to DENV infection of the four serotypes. The Huh-7 cells were infected at an MOI 1 with a strain of each DENV serotype, and at 48 hpi the viral antigen E was detected by an immunoperoxidase assay with the 4G-2 antibody. (A) Huh-7 cells not infected. (B) Specificity control without primary antibody on Huh-7 cells infected with DENV-4 414. Huh-7 cells infected with DENV-1 116 (C), DENV-2 216 (D), DENV-3 S8 (E) or with DENV-4 416 (F).
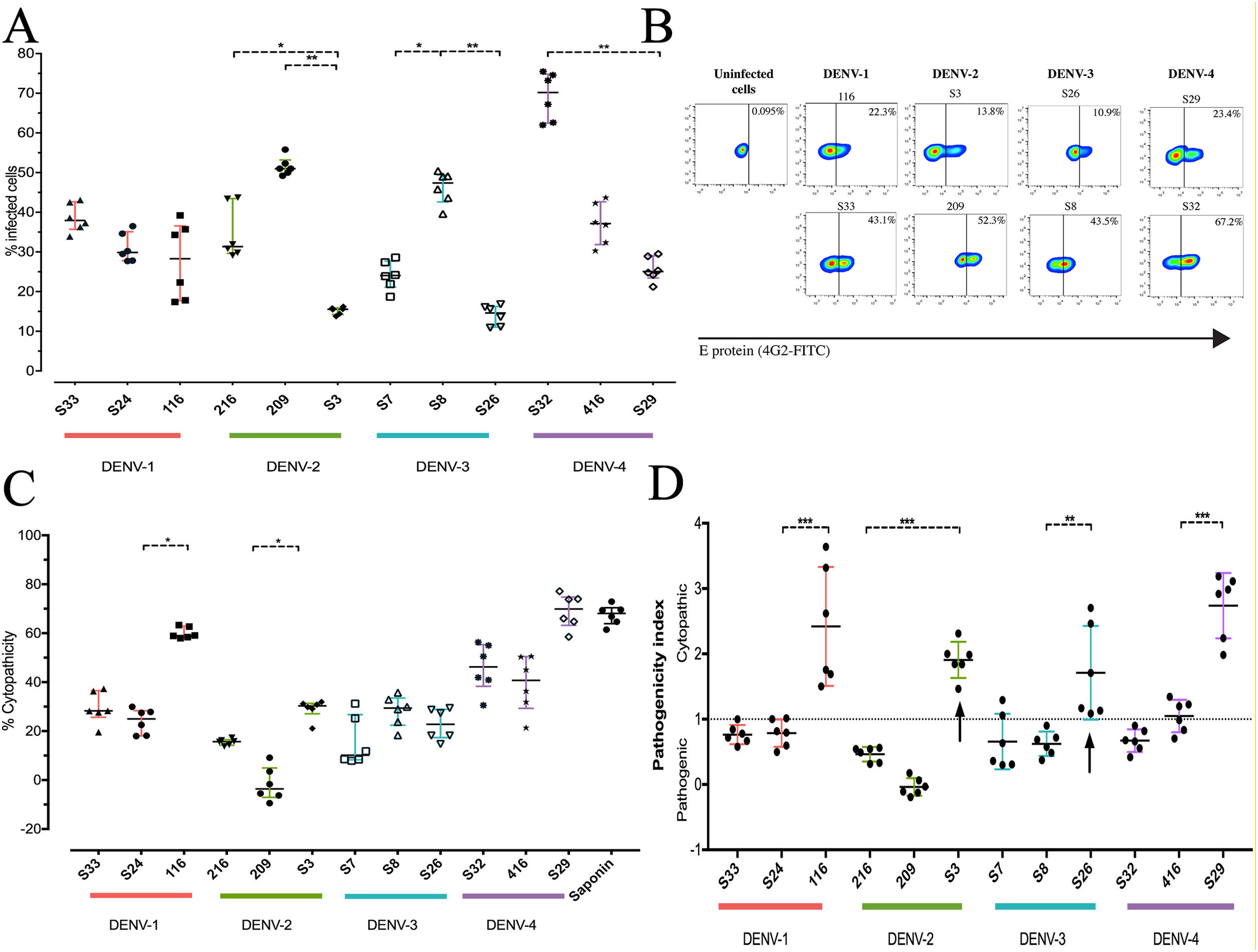
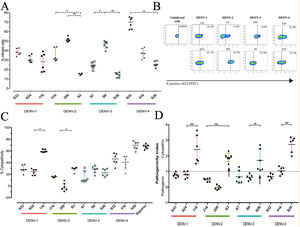
To determine the biological properties of DENV in human hepatocytes, Huh-7 cells were infected at an MOI of 0.5, and at 72h post infection (hpi), the rate of cell-cell transmission of the virus, or pathogenicity, and the effect of the infection on cell viability, or cytopathicity, were evaluated. When the percentages of infection were compared, a heterogeneous behavior was observed between strains of the same DENV serotype. The DENV-1 strains showed infection percentages between 27.8% and 38.7%, with no significant differences between the strains. On the other hand, the strains belonging to the serotypes DENV-2, DENV-3 and DENV-4 presented differences within serotypes (p<0.05): the DENV-2 strains infected between 15.21% and 51.6%, the DENV-3 strains between 14.0% and 46.3%, and infection with the DENV-4 strains reached percentages between 25.7% and 69.2% (Fig. 3 A, B). As shown in Fig. 3 C, we evaluated cell death induced by DENV infection (using the MTT assay) and found that DENV-1 and DENV-4 strains induced the highest levels of cell death (24–60% and 39.29–69.02%, respectively). The DENV-2 strains caused less cell death with values between −1.7% and 29%, while the DENV-3 strains induced 15% and 28% cell death. The variability in the capacity to induce cell death between the DENV strains shows that the viral serotype is not the only determining factor of the biological characteristics of circulating viruses.
DENV strain-dependent infection capacity and cytopathicity. The Huh-7 cells were infected with an MOI 0.5 of DENV strains, and cells were analyzed 72h post-infection. (A) Infection percentage determined to individual strain. (B) Expression of E antigen in Huh-7 cells infected with DENV of the four serotypes (two strains are shown for each serotype) by Flow cytometry using the antibody 4G2-FITC (C) Cell death percentage induced by DENV infection. (D) Pathogenic index for the 12 strains of DENV. Two independent experiments with three replies were plotted. A and C horizontal lines indicate median of IQR. D horizontal lines indicate means +/− SD. * p<0.05; ** p<0.005; *** p<0.0005.
The pathogenicity index (PI) was determined to deepen the analysis of how representative the strains included in this study are of naturally circulating populations. The flow cytometry and cell viability test values were used to calculate the index for each strain. We show that each serotype contains strains with different PI values, of which one of three strains of each serotype had a cytopathic PI (PI>1) value, while the remaining strains presented a pathogenic PI (PI<1) values (Fig. 3 D). These findings suggest that the strains belonging to each serotype of DENV present biological variability that could be a representative sample of naturally circulating strains.
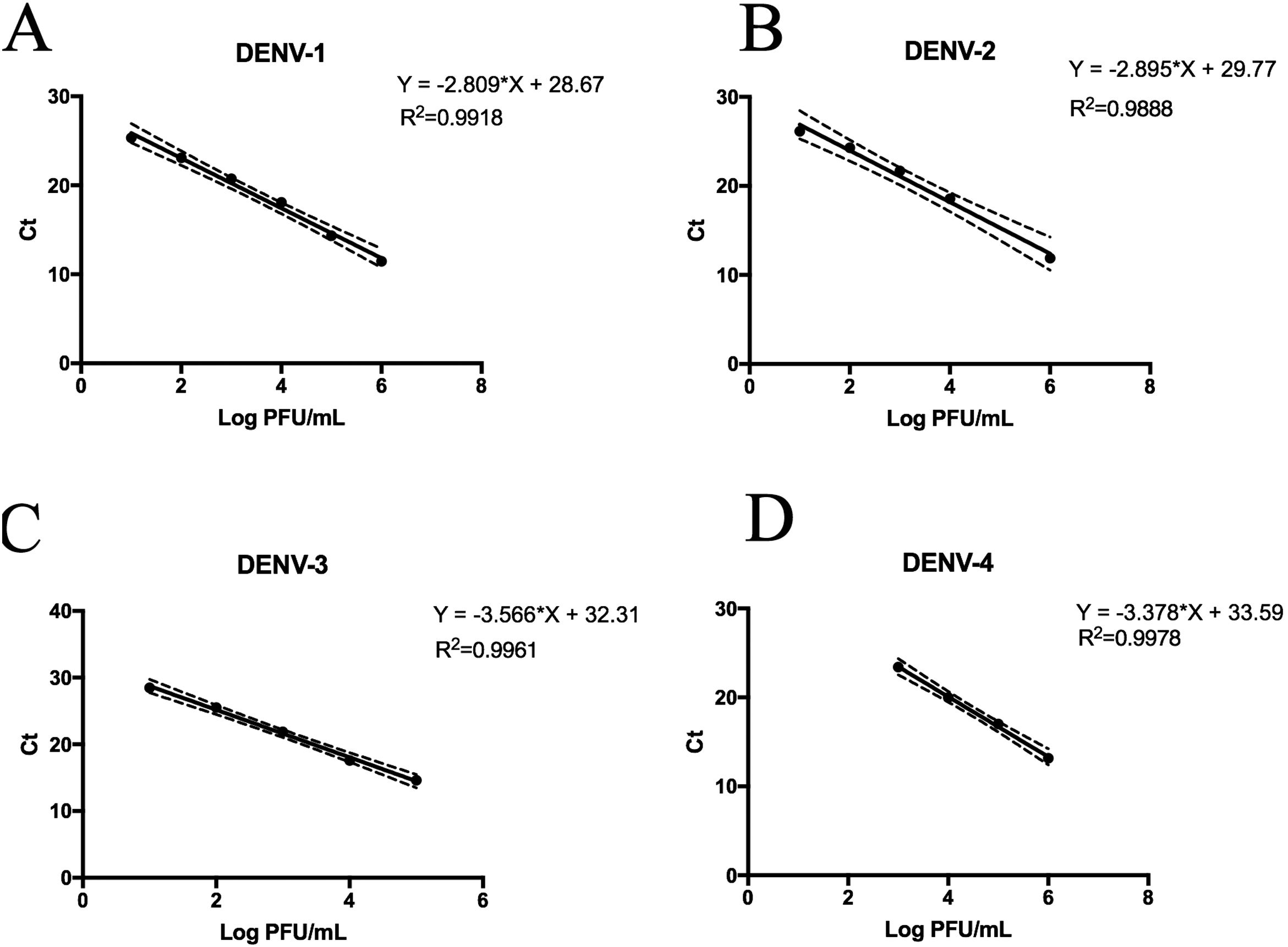
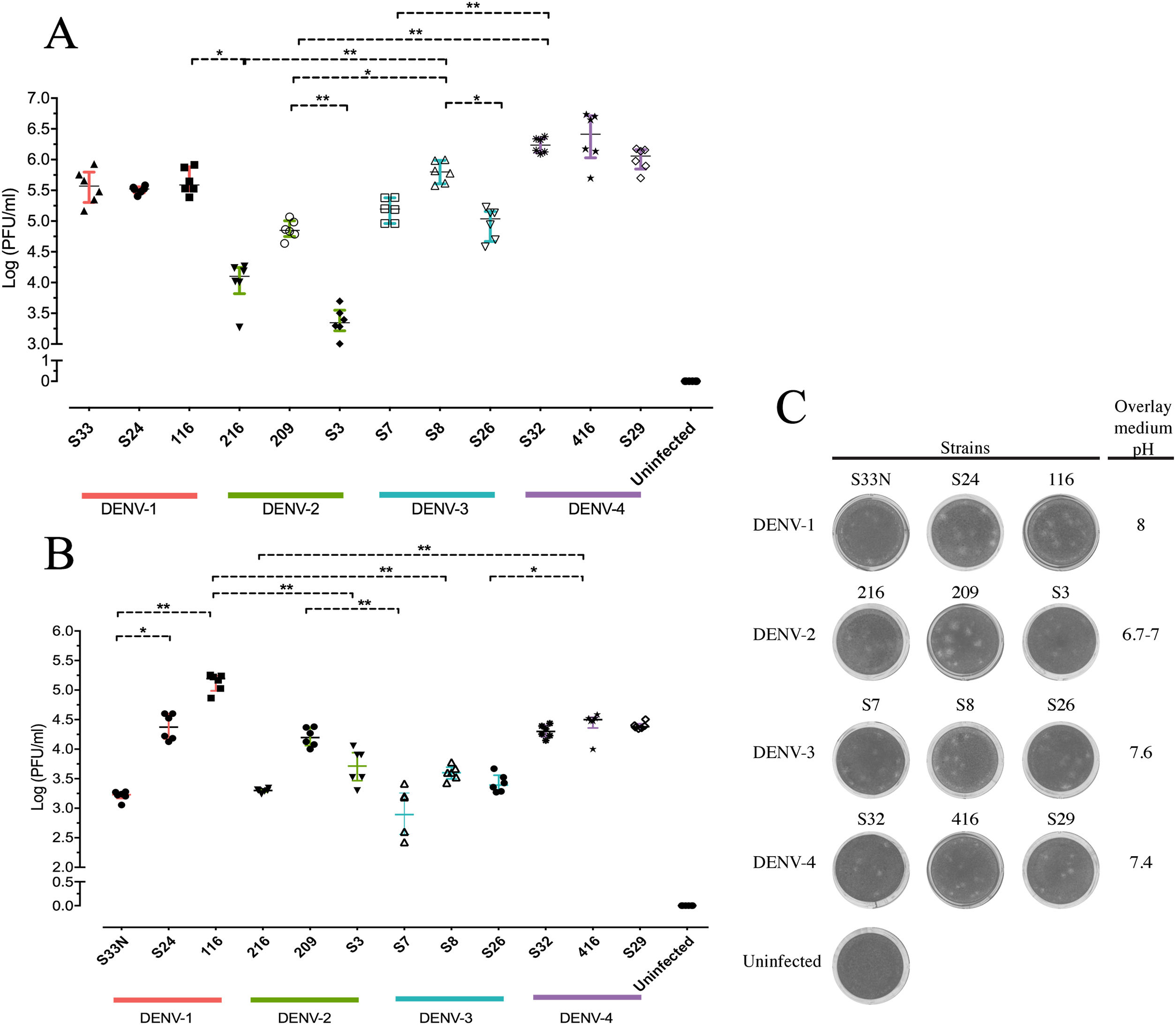
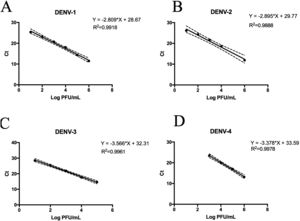
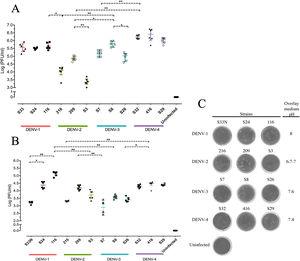
Replicative fitness characteristics are determined by the serotype and the viral strainThe two experimental approaches used for the quantification of replicative fitness, qPCR and plaque assay allowed us to determine the influence of both serotype and viral strain factors on the viral replicative fitness of DENV belonging to the four serotypes. Absolute quantification by qPCR showed that the replicative fitness could be influenced by the serotype with some variability between some strains. The standard curves for absolute quantification of replicative fitness for each viral serotype are shown in Fig. 4. The highest replicative fitness was achieved by DENV-1 and DENV-4 strains with values between 5.5 and 6.25 PFU/ml. In contrast, the lowest replicative fitness was recorded for the DENV-2 strains with 3.36–4.86 PFU/ml range, followed by DENV-3 with values between 4.95 and 5.79 PFU/ml (Fig. 5 A). The evaluation of the replicative fitness between strains of the same serotype showed significant differences between the strains belonging to DENV-2 and DENV-3. For example, the strains DENV-2 209 and DENV-2 S3 had mean values of 4.78 and 3.36 PFU/ml, respectively (p<0.05), while the strains DENV-3 S8 and DENV-3 S26 had values of 4.95 and 5.79 PFU/ml, respectively (p<0.05). For serotypes DENV-1 and DENV-4, no differences were found between the strains. These results indicate that the replicative fitness of DENV as determined by qPCR depends both on strain-specific biological characteristics and on the specific characteristics of each serotype.
Standard curves of SYBR Green RT-qPCR for the titration of four dengue virus serotypes. Serial dilution of DENV was made before RNA extraction and amplification for each serotype. (A) Standard curve for DENV-1. Dilutions from 101 to 106 PFU/ml were made from the DENV-1 116 strain. (B) Standard curves for DENV-2. Dilutions from 102 to 106 PFU/ml were made from the DENV-1 216 strain. (C) Standard curve for DENV-3. Dilutions from 101 to 105 PFU/ml were made from the DENV-1 S23 strain. (D) Standard curve for DENV-4. Dilutions from 103 to 106 PFU/ml were made from the DENV-4 416 strain. Mean values of cycle thresholds corresponding a to standard plaque forming units are plotted. Each correlation coefficient and regression equation with 95% CI are shown.
Replicative fitness characteristics are determined by the serotype and the viral strain. Huh-7 cells were infected at an MOI 0.5 with any one of four dengue virus serotypes, and the cells were analyzed 72h post-infection. (A) Replicative fitness quantification by qPCR. (B) Replicative fitness quantification by plaque assay. (C) Plaque assay with conditions of pH overlay medium and incubation days in a serotype-dependent manner (Crystal violet stain). Two independent experiments with three replicates where plotted. Horizontal lines indicate median IQR. *p<0.05; **p<0.005; ***p<0.0005.
Interestingly, after evaluation of replicative fitness by plaque assay, we again found that fitness is affected by the serotype with internal variability between strains of the same serotype. In addition, we report here that there is an optimal media pH for plaque formation for each serotype. For example, DENV-1 developed plaques at pH 8.0, DENV-2 at pH 6.9–7.0, DENV-3 at pH 7.6, and DENV-4 at pH 7.4, which could represent some specific characteristics of the viral cycle of each serotype (Fig. 5 C). Using the plaque assay approach, we found that DENV-1 and DENV-4 strains had the highest replicative fitness with values between 3.2 and 5.0 PFU/ml and 4.2–4.5 PFU/ml, respectively. In contrast, DENV-2 and DENV-3 strains had the lowest values (between 3.3 and 4.2 PFU/ml and 2.8–3.42 PFU/ml, respectively). Differences in replicative fitness between strains were only observed for DENV-1, with 5.1 PFU/ml calculated for strain 116, and 3.2 PFU/ml for strain S33, which corresponded to the upper and lower values of the range for the three strains. Although no significant differences in replicative fitness were found for the DENV-2 and DENV-3 strains (Fig. 5 B), there was an internal variation based on qPCR quantification (Fig. 5 A). These results confirm that the replicative fitness is influenced by the viral serotype, although the tendency to internal variability between the strains of each serotype is evident.
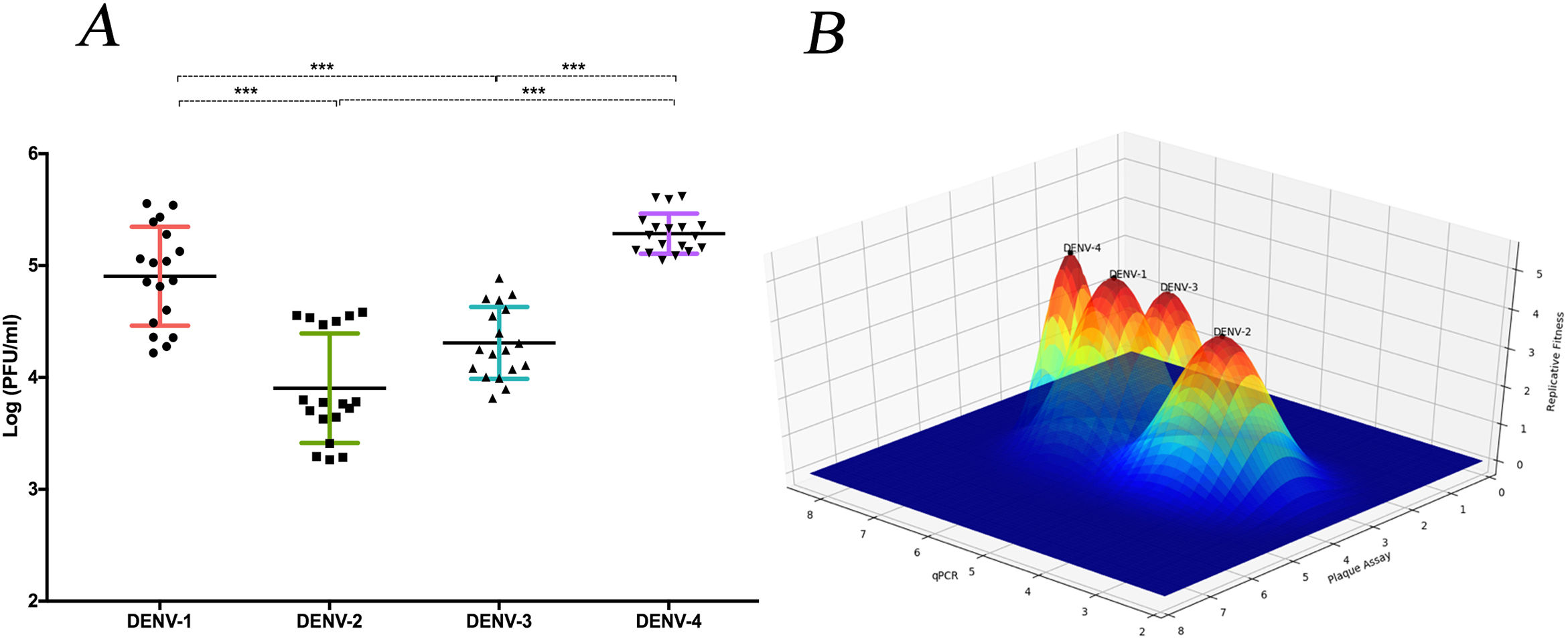
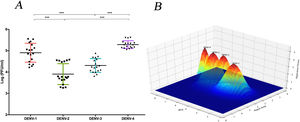
The replicative fitness average is different between DENV serotypesThe replicative fitness average of each serotype was defined as the fitness average of the three DENV strains of each of the four serotypes that were included in the study. DENV-1 and DENV-4 serotypes had the highest fitness values at 4.9 and 5.2 PFU/ml, respectively, but they were not significantly different (p>0.05). In contrast, DENV-2 and DENV-3 serotypes had the lowest fitness values, 3.9 and 4.3 PFU/ml, respectively, which were not significantly different (Fig. 6 A and B). In addition, each strain had a different fitness peak average, which demonstrates the heterogeneity and internal plasticity of the viral populations that belong to each serotype. We observed that the DENV-4 strains had a lower heterogeneity compared to the other three serotypes. The above data show that replicative fitness is influenced by the viral serotype, although there is a heterogeneous composition among the populations that comprise each type of DENV (Fig. 6 A).
The average replicative fitness in Huh7 cells is differential between dengue serotypes. (A) The Replicative fitness for each serotype was estimated using the mean values of the replicative fitness obtained by both qPCR and plaque assay. The mean values to three strains with three replicates are plotted. Horizontal lines indicate mean and SD.*p<0.05; **p<0.005; ***p<0.0005. (B) Fitness Landscape depends on the dengue serotype. The height of the pics represents the replicative fitness for each serotype (mean of replicative fitness by qPCR and plaques assay). The values showed in the “y,” and “x” axes correspond to the mean of replicative fitness by qPCR and Plaque assay, respectively.
The study of the replicative fitness in circulating viral populations and its correlation with the sustained prevalence of all serotypes in Colombia in addition to their impact on the severe manifestations of the disease, is as relevant as the efforts directed to vector control and improvement of epidemiological surveillance. Our research allowed us to establish a collection of DENV strains of each of the four serotypes isolated from the sera of patients with apparent and inapparent infection. The collection has a low number of passages in A. albopictus mosquito cells that guarantees a phenotype similar to naturally circulating viral populations. We found that the infectious capacity and the cytopathic effect induced by DENV are dependent on the viral strain and are not specifically determined by the viral serotype, which was demonstrated in the in vitro infection of Huh-7 human hepatocytes. We classified the viral strains by the pathogenicity index proposed in this study and found both cytopathic and pathogenic strains within the same viral serotype. Additionally, our report is of great scientific interest because it showed that the Colombian DENV strains have a differential replicative fitness by serotype and heterogeneous strain-dependent fitness within each serotype. This was evidenced by the serotypes DENV-1 and DENV-4 displaying the highest replicative fitness followed by DENV-2 and DENV-3.
DENV reference strains are widely used for virologic studies and have been cultured under different laboratory conditions, which results in the accumulation of genetic variations.33 These variations do not faithfully reflect the biological behavior of natural DENV populations in Colombia. On the other hand, the immune response and the history of viral infections of the patient have been described as factors that influence the population dynamics of DENV. The analysis of variations in viral proteins from patients with secondary DENV infection showed unique variants that differ from those found in patients with a primary infection. These variations were also described in protein E, one of the most immunogenic proteins to which the immune response mediated by antibodies is directed.34In vitro tests show that a DENV population exposed to an "anti-fusion loop" antibody targeting the E protein results in the selection of variants with mutations in this region, which allow new populations to evade the neutralizing effect of this antibody, guaranteeing subsistence of the virus.35
Evidence shows that population immunity exerts selection pressure on DENV, which explains the heterogeneity in replicative fitness reported between strains from different regions.36 Colombia is a country with exceptional epidemiological dynamics due to the simultaneous circulation of the four serotypes of DENV, which favors successive infections with different viral serotypes throughout an individual’s life. This epidemiological behavior leads to a DENV selection that results in viral populations that are unique and not homologous to DENV strains originating in other countries. Our study provides robust knowledge on twelve representative strains of the four DENV serotypes that are very useful for the investigation of local epidemiological dynamics.
In this study, we showed that the biological characteristics, such as infection capacity and effect of infection on cell viability, are influenced by the strain and not by the viral serotype of DENV. Previous studies have found biological characteristics of DENV that were dependent on the viral strain, such as the ability to regulate the type I interferon response,37 as well as differential NS1 protein secretion rates between strains of the same serotype.38 Based on the variable biological behavior between viral strains, we propose an index of pathogenicity (PI), which allows classification of the viral strains based on the phenotypic characteristics that represent critical events in the viral cycle. The PI of the DENV strains in this study showed that the DENV-2 strains are the most phenotypically heterogeneous, followed by the DENV-4, DENV-3, and DENV-1 strains, which means that pathogenic and cytopathic strains exist within each serotype. The phenotypic diversity found among the local DENV strains of the same serotype demonstrates the complex picture that will have to be addressed for the study of viral populations in endemic regions.
According to our results, the viral serotype influences the replicative fitness of the Colombian strains of the four DENV serotypes. Previously, it was shown that the ability of the viral serotype to form lytic plates depended on the pH of the overlay medium used in the plating assay.39 The four serotypes used in this study exhibited the same pH dependence since the DENV-1 and DENV-3 strains needed basic medium overlay, while the DENV-2 and DENV-4 strains needed acidic medium overlay for the optimal development of lytic plaques during infection. This finding could be related to the differential translocation of the NS5 protein in response to the extracellular pH since the NS5 protein of DENV-2 predominantly localizes to the nucleus at a pH of 6.0, while at pH 8.5, it is located in the cytoplasm.40 Additionally, the NS5 viral polymerase of DENV-2 and DENV-3 predominantly localizes to the nucleus, while that of DENV-1 and DENV-4 is localized to the cytoplasm due to the absence of a nuclear localization sequences (NLS) in the NS5 viral polymerase of these serotypes.41 Likewise, the subcellular localization of the NS5 protein in DENV serotypes has been associated with differences in the production of viral progeny since mutations in the NLS sequences that inhibit the translocation of the protein to the nucleus cause a reduction in viral progeny production.42 The cellular localization of the NS5 protein described above could explain the differential replicative abilities observed in this study, in which we found that strains grouped by viral serotype: the DENV-1 and DENV-4 strains are grouped together with the highest replicative fitness, and the DENV-2 and DENV-3 strains are grouped together by a lower replicative fitness.
The influence of the viral serotype on replicative fitness could also be explained by differences in the interaction of the virus with cellular receptors or the trafficking and release of viral progeny. The differential use of receptors among the four DENV serotypes has been previously reported. The interaction between the viral binding protein (viral attachment protein) and the expression levels of receptors in the cell membrane on the surface of the target cell affect the entry of viral serotypes. For example, polyclonal antibodies against laminin decreased DENV-1 production, whereas the viral progeny of DENV-2, DENV-3, and DENV-4 was not affected, which suggests a differential interaction between viral serotypes and cellular receptors.43 Regarding the assembly of the intracellular virus, the interaction of the N-terminus of the preM protein with the receptors KDELR1 and KDELR-2 has been reported, which is crucial for the transport of serotypes DENV-1, -2, and -3 but not for the assembly of DENV-4. Thus, the inhibition of KDEL receptor expression blocks the release of the viruses of the first serotypes, which leads to the accumulation of viral particles and deterioration of the maturation process.44 The use of more than one receptor or the availability of receptors in cell lines used in the viral infection experiments could influence the efficiency of the maturation and export of viral progeny, affecting the replicative viral fitness of the DENV serotypes. In this work, we found differences between serotypes in terms of the ability to produce viral progeny, and it is probable that DENV serotypes have evolved to use different components of the cellular machinery in the process of viral replication. In conclusion, the evidence shows that the DENV serotypes could have a differential replicative fitness, which could be linked to the expression of receptors, as well as to the cellular localization of viral proteins.
Replicative fitness is one of several factors that together could explain the hyperendemicity of DENV in Colombia. First, it is possible that the variability in replicative fitness within viral populations of the same serotype results in greater evolutionary plasticity, which allows the coexistence of strains with variable replicative fitness of the same serotype in different geographical regions. This suggests that the variability between strains determines the prevalence of one serotype by region. Second, genetic flow events from endemic countries such as Venezuela,45 which increase the genetic and phenotypic variability of populations, would prevent the establishment of one serotype. The third important factor is the high probability of secondary dengue infections caused by the antibody-dependent enhancement (ADE) mechanism, which would allow a strain with a low or high replicative fitness to persist in the population without direct competition.46 Finally, since it has been reported that 84% of dengue infections in endemic countries are mono infections and there are coinfections at a lower percentage,47 the probability of competition between strains during natural infection is low. The combination of these factors could explain the presence of multiple peaks of replicative fitness in populations that comprise a viral serotype and that persist dynamically over time and in endemic countries such as Colombia.
We report a differential replicative fitness between local serotypes circulating in our country. Based on our results, it would be necessary to evaluate how the serotype influences the immune response induced by DENV. For this evaluation, the use of established and characterized local strains and an optimal cell culture system that is permissive to infection are critical. This system would allow tracing the kinetics of expression and production of multiple inflammatory cytokines. Additionally, it could be possible to study the correlation between the infectious viral serotype and the clinical outcome of dengue.
The DENV serotypes most frequently associated with severe clinical symptoms in South America are DENV-2 and DENV-3, followed by DENV-1 and DENV-4.15,16 Since we found a high replicative fitness for DENV-1 and -4, it is possible that these serotypes do not induce an exacerbated inflammatory response and thus maintain an interaction with the host cell that favors the production of viral progeny and leads to a highly efficient viral replication. In contrast, strains belonging to serotypes DENV-2 and DENV-3 with lower replicative fitness could induce an aggressive inflammatory response, which could diminish the replicative fitness of the virus and at the same time increase the probability of a severe clinical outcome directed by an out-of-control immune response, previously described as a cytokine storm.48
The collection of strains of the four DENV serotypes established and characterized in this study may be used in future research on the pathogenesis of severe manifestations of DENV infection in humans and the role of strains and serotypes in the establishment of circulating serotypes in endemic areas.
ConclusionsThe differential replicative fitness between serotypes observed in Colombian DENV strains isolated and characterized genotypically and phenotypically in this study reflects the heterogeneity of locally circulating populations and contributes to the understanding of the complex physiopathology of the disease in infections caused by the different serotypes. However, we found that the infectious capacity and the cytopathic effect are dependent on the viral strain, which demonstrates that some biological characteristics of the viruses are not shared within the same serotype and could contribute, together with specific characteristics of the host, to the severity of the clinical outcome. The cell culture model of human hepatocytes Huh-7 is optimal for viral replication infection studies because it allows efficient production of the viral progeny by the four DENV serotypes. The virus collection of strains of the four DENV serotypes established and characterized in this study can be used in future research on the pathogenesis of severe manifestations of DENV infection in humans.
Conflicts of interestThis manuscript has not been published and is not under consideration for publication elsewhere. We have no conflicts of interest to disclose. This study was funded by the Vicerrectoría de Investigaciones-Universidad El Bosque (Project PCI-2015-8327).