Indiscriminate use of antimicrobials has caused the worldwide emergence of multidrug resistant bacteria (MDR). Glucose non-fermenting Gram-negative bacilli (NF-GNB) that show resistance to several antimicrobials, including colistin, have been reported. The highest rates of this type of resistance have been registered in Asia and Europe.1 This fact has created problems in therapy, since the colistin is used as last alternative to treat infections caused by MDR bacteria.2Pseudomonas aeruginosa is a skilful microorganism that causes severe opportunistic infections in immunocompromised patients due to its metabolic versatility.3 Isolation of P. aeruginosa resistant to almost all available antimicrobials is increasing in Intensive Care Units (ICU), leading to an increase in the number of healthcare-associated infections (HAI), significantly influencing morbidity, mortality, and high costs of treatment of infected patients.1
In a prospective study developed between August 2015 and January 2016 we isolated 293 NF-GNB of various clinical specimens from patients admitted to a university hospital in the central region of Rio Grande do Sul, Brazil. Identification and antimicrobials sensitivity test were performed through the automated system VITEK®2 (bioMérieux). Data were interpreted according to the document M100-S25 of the Clinical and Laboratory Standards Institute [CLSI, 2015], considering resistant the isolated specimens which showed intermediate or resistant colistin profiles.
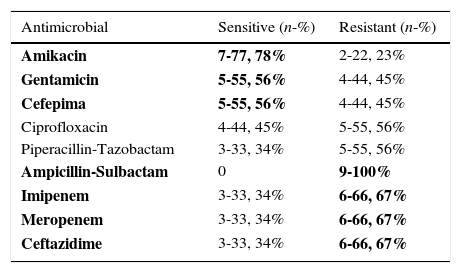
From the 293 NF-GNB assessed in this study, only two genera were identified: 78.83% (231) P. aeruginosa and 21.16% (62) Acinetobacter spp. Colistin resistance was identified in nine isolated strains of P. aeruginosa (9/231-3,89%) and none in Acinetobacter spp. The predominant age group of patients in whom colistin-resistant strains were identified was 41–69 years old (77.77%); this age-range also showed expressive resistance to carbapenems (66.67%) (Table 1).
Sensitivity profile of 9 P. aeruginosa isolates resistant to colistin.
| Antimicrobial | Sensitive (n-%) | Resistant (n-%) |
|---|---|---|
| Amikacin | 7-77, 78% | 2-22, 23% |
| Gentamicin | 5-55, 56% | 4-44, 45% |
| Cefepima | 5-55, 56% | 4-44, 45% |
| Ciprofloxacin | 4-44, 45% | 5-55, 56% |
| Piperacillin-Tazobactam | 3-33, 34% | 5-55, 56% |
| Ampicillin-Sulbactam | 0 | 9-100% |
| Imipenem | 3-33, 34% | 6-66, 67% |
| Meropenem | 3-33, 34% | 6-66, 67% |
| Ceftazidime | 3-33, 34% | 6-66, 67% |
Colistin-resistant P. aeruginosa were predominantly isolated from clinical specimens obtained from soft tissues (3/9-33.33%) and respiratory (sputum and tracheal secretion – 2/9-22.22%) secretions. The hospital wards in which resistant strains were initially identified were internal medicine (2/9-22.23%), surgery (2/9-22.23%), recovery room (2/9-22.23%), nephrology, adult ER, and ICU (1/9-11.11% each).
From the nine patients in whom colistin-resistant P. Aeruginosa was isolated, three progressed to death due to septicemia, four were oncologic patients under antineoplastic and radiotherapy regimens, one post-kidney transplant patient, two diabetes mellitus patients, and two chronic smokers/alcoholics. Therefore, we ratify that patients’ immunodepression make them vulnerable to infections caused by colistin-resistant microorganisms.
The extensive use of colistin in veterinary medicine to treat gastrointestinal infections caused by Enterobacteriaceae has contributed to chromosomal mutations mediated by the plasmid mcr-1 to codify resistance to colistin in Gram-negative bacilli. Fernandes and colleagues reported on the evidence of mcr-1 originated from Escherichia coli isolated from food of animal origin in Brazil since 2012.4 This silent dissemination of E. coli resistance mediated by this plasmid is having an impact in Brazilian ICUs. The emergence and dissemination of MDR bacteria associated with high rates of treatment failure have fostered the use of poliximins, the main therapeutic option for the treatment of MDR infections, especially those caused by P. aeruginosa, Acinetobacter baumannii, and Klebsiella pneumoniae, producers of SPM-1, OXA-23 or KPC-2.5 Additional studies are being developed by our research group to identify the gene mcr-1 in isolated strains that show resistance to colistin.
Conflicts of interestThe authors declare no conflicts of interest.