Detection of drug resistance plays a crucial role in tuberculosis (TB) treatment and prevention of Mycobacterium tuberculosis (MTB) transmission. The aim of this study was to determine the levels and patterns of resistance of MTB isolates to two key anti-TB drugs (rifampicin, RIF and isoniazid, INH) and the type of mutations in drug resistance genes (rpoB, katG and inhA) of the isolates at the southern coastal region of Andhra Pradesh, India, using commercially available GenoType MTBDRplus assay under the Revised National TB Control Program.
MethodsGenoType MTBDRplus assay was performed on 2859 sputum smear-positive samples and the mutations in the genes responsible for resistance (rpoB, katG and inhA) were analyzed.
ResultsAmong the line probe assay (LPA) valid isolates (2894), 1990 (68.76%) were drug susceptible, 437 (15.13%) were INH monoresistant, 104 (3.59%) were RIF monoresistant, and 363 (12.54%) were multidrug resistant. Codon 531 of rpoB gene and codon 315 of katG gene were found to have the highest mutation frequency for RIF resistance (270/467; 57.81%) and INH resistance (501/800; 62.62%), respectively. The RIF resistant rpoB mutations observed in the samples were S531L (57.81%), H526Y (8.56%), D516V (6.42%), and H526D (6.20%). Mutations in inhA promoter were found in 24.75% INH resistant isolates with C15T being the most common (85.85%). The turnaround times of the LPA test were from 48 to72h.
ConclusionThe frequency of mutations in MTB in the coastal region of Andhra Pradesh, India, is similar to that in retreatment cases from most settings, with close to 80% in rpoB codon 516, 526, and 531, and over 80% in codons katG 315 and/or inhA promoter. The increase in INH monoresistance underlines the need for greater enforcement of national TB control programs.
Tuberculosis (TB), caused by Mycobacterium tuberculosis (MTB), is one of the leading causes of death worldwide. The most recent reports of World Health Organization (WHO) show that there were 9.0 million new cases of TB and 1.5 million deaths due to TB.1 Despite the evidence that TB is slowly declining, worldwide emergence and spread of multidrug resistance (MDR; resistant to at least one of the key first-line anti-TB drugs (isoniazid (INH) and rifampicin (RIF)) and extensive drug resistance (XDR) (MDR plus additional resistance to a fluoroquinolone and any second-line injectable drug) in MTB strains have become a major obstacle for TB control.2
The emergence of MDR and XDR TB has left clinicians with hardly any treatment options, and their spread has created a serious threat to public health. MDR and XDR TB are on the rise mainly due to inappropriate drug regimen, patient defaulting, previous anti-TB treatment, delay in diagnosis and initiation of effective treatment, and primary infection with MDR TB strains. Patients with MDR and XDR TB (those who do not respond to treatment) can be a constant source of transmission of drug-resistant MTB. Accurate and rapid detection of MDR TB is critical for decreasing the transmission of TB and reducing the amplification of drug resistance.3
Acquisition of drug resistance in MTB does not occur as a result of horizontal transfer of resistance-determining genes or region, but results from mutations (caused by substitutions or deletions or insertions of nucleotide sequences) in specific resistance-determining regions of the gene targets or their promoters or activating enzymes of anti-TB chemotherapeutic agents. Mutations have been reported in the genes katG, inhA and ahpC (resistance against INH); rpoB (resistance against Rifampicin, RIF); and inhA (resistance against Ethionamide).4
Conventional culture and drug susceptibility testing (DST) for MTB on solid/automated liquid culture systems is a time-consuming procedure (takes up to 8–12 weeks), suffers from higher contamination rates, and requires considerable equipment, media, and technical expertise or human resources.5 However, based on the knowledge of reported mutations of the anti-TB drug resistance genes, rapid molecular diagnostic line probe assay (LPA) has been developed for the detection of drug resistance in MTB. INNO-LiPA RifTB (Innogenetics N.V, Ghent, Belgium), GenoType®MTBDRplus (for detection of INH and RIF resistance), and GenoType®MTBDRsl (for detection of resistance against ethambutol, fluoroquinolones and aminoglycosides) (Hain Lifescience, Nehren, Germany) are commercially available types of LPA that are being used to detect simultaneously the reported mutations in the drug resistance genes of MTB. In 2008, genotype MTBDRplus LPA has been recommended for rapid detection of drug resistance of MTB by WHO.1
Many studies have shown that the specificity and sensitivity of GenoType MTBDRplus assay for RIF and INH resistance are found to be comparable to conventional phenotypic drug susceptibility testing (DST).6–8 A meta-analysis study carried out in 2008 found that the GenoType MTBDRplus assay has a sensitivity of 98% for detecting RIF resistance and 89% for detecting INH resistance, and specificity of 99% for both RIF and INH.9
Globally, many reports have been published on detection and diagnosis that are based on rapid advanced molecular assays. However, owing to the geographical diversity of MTB clinical isolates across the world, there is a possibility of varying efficacy of molecular genotyping methods, which may have diagnostic implications. In addition, the mutations responsible for resistance could differ with region and thus provide an insight into the epidemiology of the disease. However, the magnitude of drug resistant MTB isolates and pattern of drug resistance mutations is not well known in the south coastal region of Andhra Pradesh, India. Therefore, the present study was undertaken to determine the anti-TB drug resistance and associated patterns of mutations in rpoB, katG and inhA genes of MTB isolates from patients of this region using commercially available GenoType MTBDRplus assay under the Revised National TB Control Program (RNTCP) of India, and to evaluate the possible risk factors associated with mycobacterial resistance in the patients of this region.
Materials and methodsMycobacterium tuberculosis isolates, study sites, and sample processingThis study was part of a programmatic management of drug resistant TB, conducted by Damien Foundation India Trust, with the support of the District TB Control Officer (DTCO), Nellore, Andhra Pradesh, India. This center has a TB culture and drug susceptibility testing (C & DST) referral laboratory for RNTCP, accredited by the National Mycobacteriology Accreditation System of Central TB Division Ministry of Health, Government of India. Data in this study are from patients who had registered during a period of one year.
Five thousand and eighty-seven non-duplicate (one isolate per patient) clinical isolates of MTB were obtained from the clinical samples of pulmonary TB patients from microscopy centers operating under the aegis of the RNTCP, located in Southern districts of Andhra Pradesh (Ananthapur, Chittoor, Prakasam, Kadapa, Kurnool, and Nellore), India. Standard microbiological techniques were used for isolation and identification of the isolates. All the chosen cases were pulmonary TB patients with symptoms of weight loss, chest pain, night fever, blood mixed sputum, and so forth. Information about patients was obtained by physicians on a standard form that is used for patients suspected to have TB. This study received ethical clearance from the ethics committee of Krishna Institute of Medical Sciences (KIMS), Nellore. Patients were identified and managed according to RNTCP guidelines (RNTCP, 2009 guidelines). Informed consent was obtained from all participants included in the study. According to RNTCP guidelines, occurrences of TB in patients who underwent category II treatment (2(SHRZE)3+1(HRZE)3 5(HRE)3) with four-month follow up were considered as previously treated cases and those in patients who underwent category I treatment (2(HRZE)3 4(HR)3) were considered as new cases (H=isoniazid, R=rifampicin, Z=pyrazinamide, E=ethambutol, S=streptomycin).
Isolation and identification of MTB was carried out in a certified microbiology laboratory using Ziehl–Neelsen (ZN) staining or acid-fast bacilli (AFB) staining technique.10 The smears were graded according to the number of bacilli seen on the slide, as per RNTCP guidelines. All smear positive (>1+AFB) sputum samples received within 48–72h of sample collection in cold chain were subjected to only LPA. Smear negative and scanty acid fast bacterial samples were subjected to culture on Lowenstein–Jensen (L–J) solid media for growth of MTB. The culture-positive samples for MTB were further subjected to LPA analysis. The samples were processed by N-acetyl-L-cysteine-sodium hydroxide (NALC-NaOH) method of digestion and decontamination.11
Line probe assay/Genotype MTBDRplus assayThe GenoType MTBDRplus assay version 2.0 (Hain Life Sciences, Nehran, Germany) was performed in accordance with the manufacturer’s instructions (http://www.hainlifescience.de). The LPA is a multiplex polymerase chain reaction (PCR) based genotypic test that identifies MTB complex and simultaneously detects the mutations that confer resistance to RIF and INH. The assay has an additional advantage over other LPAs because the GenoType MTBDRplus assay identifies mutations in the rpoB gene (coding for the ß-subunit of the RNA polymerase) for detection of RIF resistance, mutations in the katG gene (coding for the catalase peroxidase) for high-level INH resistance, and mutations in the promoter region of inhA gene (coding for the NADH enoyl ACP reductase) for low-level INH resistance. H37 RV was used as the positive control.
Identification of non-tuberculosis mycobacteria (NTM)Identification of non-tuberculosis mycobacteria (NTM) was done based on colony morphology, pigmentation, growth rate on conventional solid, and media containing p-nitrobenzoic acid (PNB). In addition, biochemical tests such as niacin test and heat-stable catalase test (pH 6.8, 68°C) were performed as per the protocols in Training Manual for Mycobacterium tuberculosis Culture & Drug susceptibility testing.10
Turnaround time (TAT)Turnaround time was calculated as the time between the date of receiving sputum sample at the primary health center and date of MTBDRplus test results.
Statistical methodsSPSS software20.0 was used for data analysis. The statistical analysis was performed using SPSS software (Version 17.0). All values are expressed in the form of percentages and the Chi-square test was applied wherever necessary. Statistical significance was set at p≤0.05.
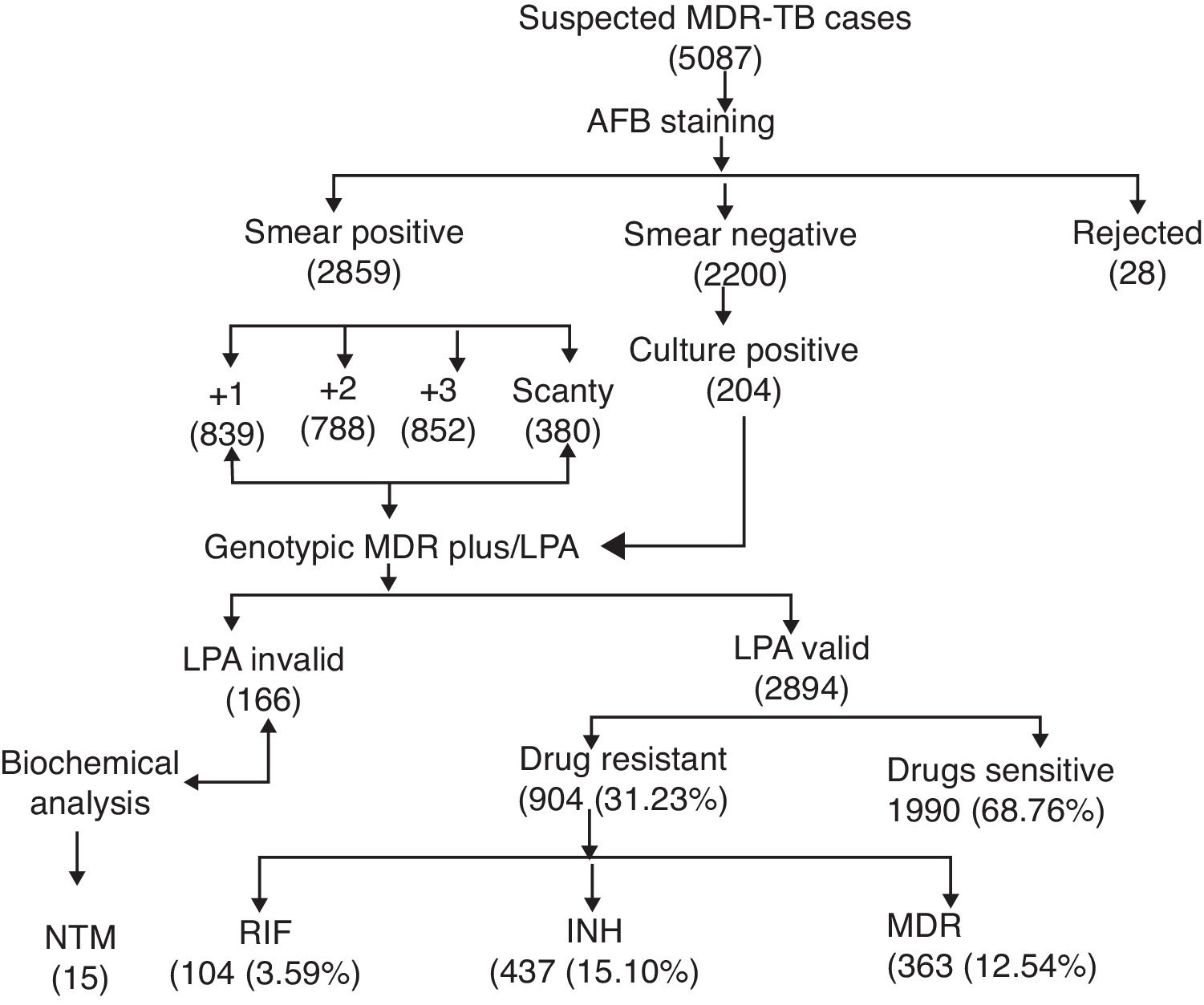
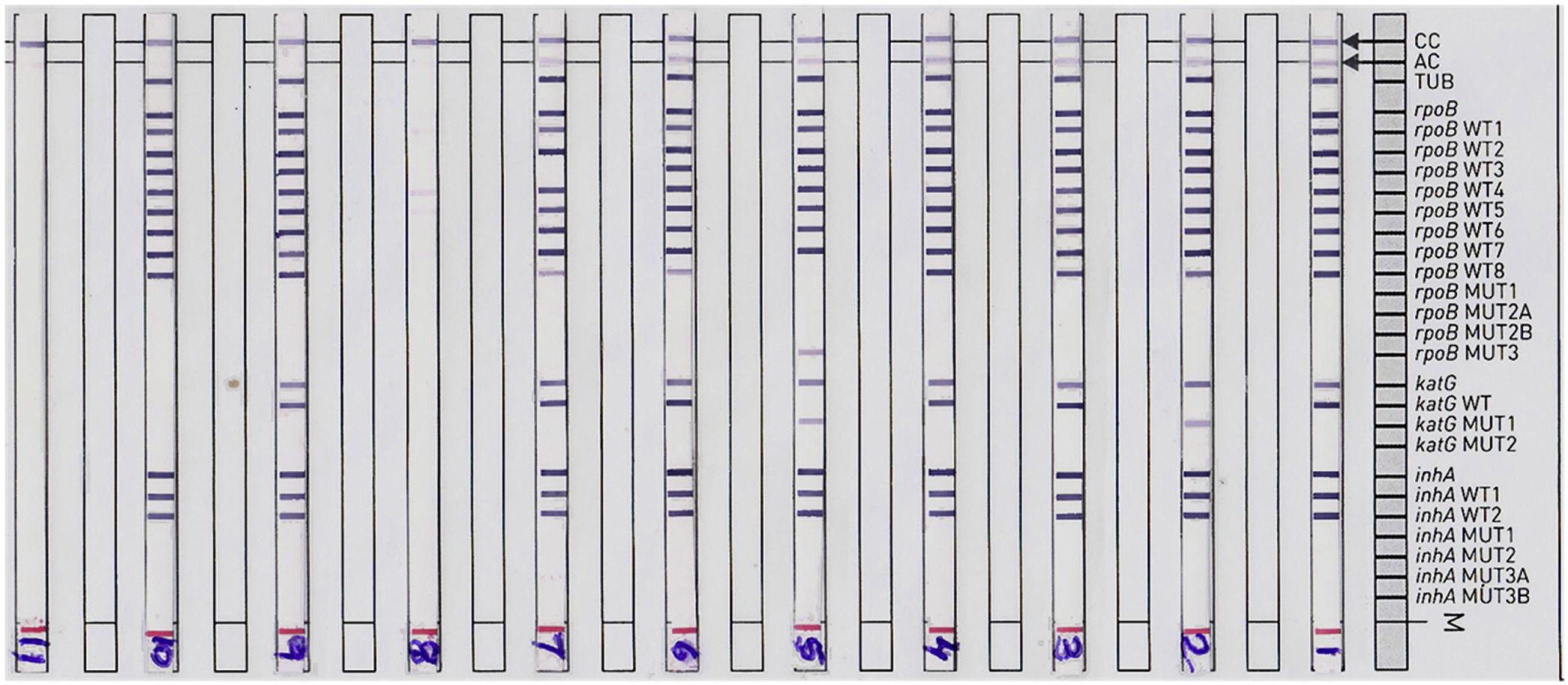
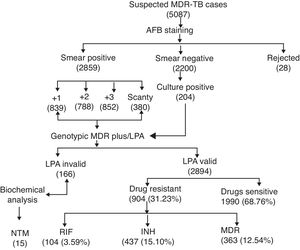
ResultsA total of 5087 sputum samples were obtained from primary health centers of various southern districts of Andhra Pradesh, India, from December 2013 to December 2014 (one-year period). Among the study isolates, 2859 (56.14%) samples were AFB smear positive and 2200 (43.24%) samples were smear negative. The smear-positive samples were graded on a scale from 0 to 3+. Three hundred and eighty (13.09%) samples gave scanty positive results and 30 samples were rejected. A total of 839 (29.34%) samples had an AFB count of one to 10 per 100 fields (smear 1+), 788 (27.56%) samples had one to nine bacilli per field (smear 2+), and 852 (29.80%) samples had more than nine bacilli per field (smear 3+) (Fig. 1). Of the smear-negative specimens, 204/2200 (9.27%) were culture positive for AFB and were included for the final LPA analysis, of which 204/204 (100%) confirmed to be MTB complex (i.e., presence of TUB band (Fig. 2)).
Representative DNA strip patterns of GenoType MTBDR-plus strip. Lane 1, M. tuberculosis complex H37Rv laboratory control strain (rpoB, katG, inhA WT); Lane 3, 4, 6, 9 & 10sensitive to Rifampicin (RIF) and Isoniazid (INH); Lane 2, INH monoresistant (katG S315T1 mutation) Lane 5, MDR-TB (rpoBS531L andkatGS315T1 mutation); Lane 7, RIF monoresistant (rpoB unknown mutation); Lane 8, TUB band absent (LPA invalid) Lane 11, DNA negative control.
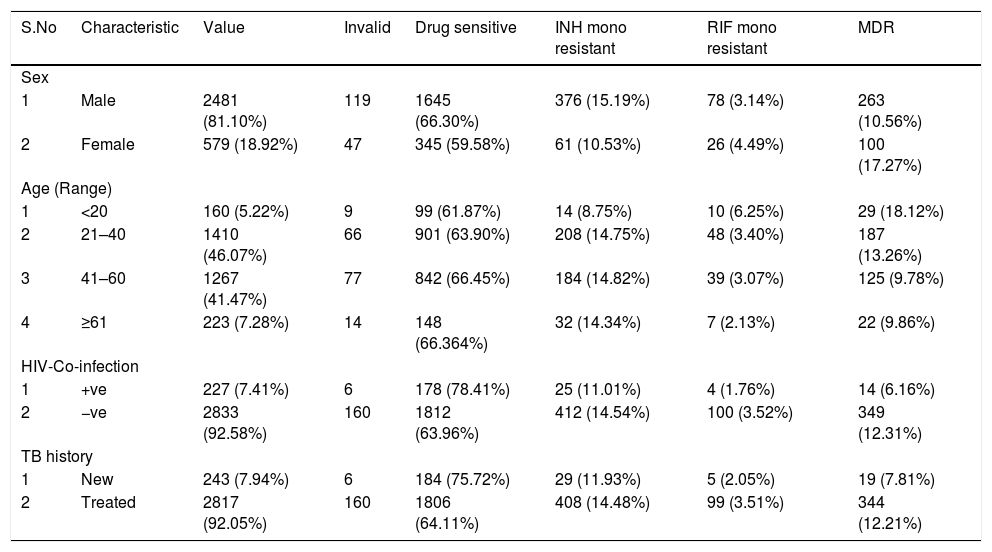
Of the final 3060 sputum samples, 2481 (81.10%) were from male patients and 579 (18.92%) from female patients. Majority of the patients were in the 21–40 (1410/3060; 46.07%; economically productive group) and 41–60 (1267; 41.47%) age groups. Out of 3060 patients, 227 (7.41%) were coinfected with HIV and MTB (Table 1). The mean age of the study patients was 41.5 years. Among the 3060 cases, 243 (7.94%) were new and 2817 (92.05%) included patients who were undergoing TB treatment or previously treated (i.e., DOTSplus category II treatment).
Distribution of host and microbial characteristics among drug-sensitive, drug-resistant, and MDR TB cases among patients of southern coastal region of Andhra Pradesh, India.
| S.No | Characteristic | Value | Invalid | Drug sensitive | INH mono resistant | RIF mono resistant | MDR |
|---|---|---|---|---|---|---|---|
| Sex | |||||||
| 1 | Male | 2481 (81.10%) | 119 | 1645 (66.30%) | 376 (15.19%) | 78 (3.14%) | 263 (10.56%) |
| 2 | Female | 579 (18.92%) | 47 | 345 (59.58%) | 61 (10.53%) | 26 (4.49%) | 100 (17.27%) |
| Age (Range) | |||||||
| 1 | <20 | 160 (5.22%) | 9 | 99 (61.87%) | 14 (8.75%) | 10 (6.25%) | 29 (18.12%) |
| 2 | 21–40 | 1410 (46.07%) | 66 | 901 (63.90%) | 208 (14.75%) | 48 (3.40%) | 187 (13.26%) |
| 3 | 41–60 | 1267 (41.47%) | 77 | 842 (66.45%) | 184 (14.82%) | 39 (3.07%) | 125 (9.78%) |
| 4 | ≥61 | 223 (7.28%) | 14 | 148 (66.364%) | 32 (14.34%) | 7 (2.13%) | 22 (9.86%) |
| HIV-Co-infection | |||||||
| 1 | +ve | 227 (7.41%) | 6 | 178 (78.41%) | 25 (11.01%) | 4 (1.76%) | 14 (6.16%) |
| 2 | −ve | 2833 (92.58%) | 160 | 1812 (63.96%) | 412 (14.54%) | 100 (3.52%) | 349 (12.31%) |
| TB history | |||||||
| 1 | New | 243 (7.94%) | 6 | 184 (75.72%) | 29 (11.93%) | 5 (2.05%) | 19 (7.81%) |
| 2 | Treated | 2817 (92.05%) | 160 | 1806 (64.11%) | 408 (14.48%) | 99 (3.51%) | 344 (12.21%) |
INH, Isoniazid; RIF, Rifampicin; MDR, Multidrug Resistant; Invalid-Samples were either negative or results were not clear.
The MTBDRplus test was done for 3060 TB suspected samples and among these, valid results were obtained for 2894 (94.57%) samples. The remaining 166 (5.42%) samples were either negative or results were not clear; hence, these isolates were not included for further MTBDRplus studies. LPA invalid samples were further subjected to bacterial culture and biochemical methods to characterize the smear-positive and LPA-negative isolates. Out of 166 acid fast isolates, 15 (0.5%) were identified as NTM based on their growth on media containing PNB, negative response to niacin test, and positive response to catalase activity at 68°C and pH 6.8. The MTBDRplus LPA invalid rate increased as 13/852 (1.5%), 78/839 (9.3%), 14/788 (1.8%), and 61/380 (16.0%); and the AFB smear grading decreased as 3+, 2+, 1+, and scanty. This correlation was statistically significant (p<0.005).
Direct MTBDRplus test results showed that 2894 sputum samples were MTB positive, that is, presence of TUB band, which bound the amplicons to the MTB complex. Drug resistance to one or more anti-TB drugs was found in 904 (31.23%) MTB isolates of 2894 patients (Fig. 1). As shown in Table 1, among MTBDRplus performed, 1990 (68.76%) isolates were susceptible to both RIF and INH, 104 (03.59%) were RIF monoresistant, 437 (15.10%) were INH monoresistant. and 363 (12.54%) were MDR (resistant to both the first line of drugs: RIF and INH) (Table 1 and Fig. 1). An average turnaround time, including specimen transportation time for LPA performed from different regions, was five days.
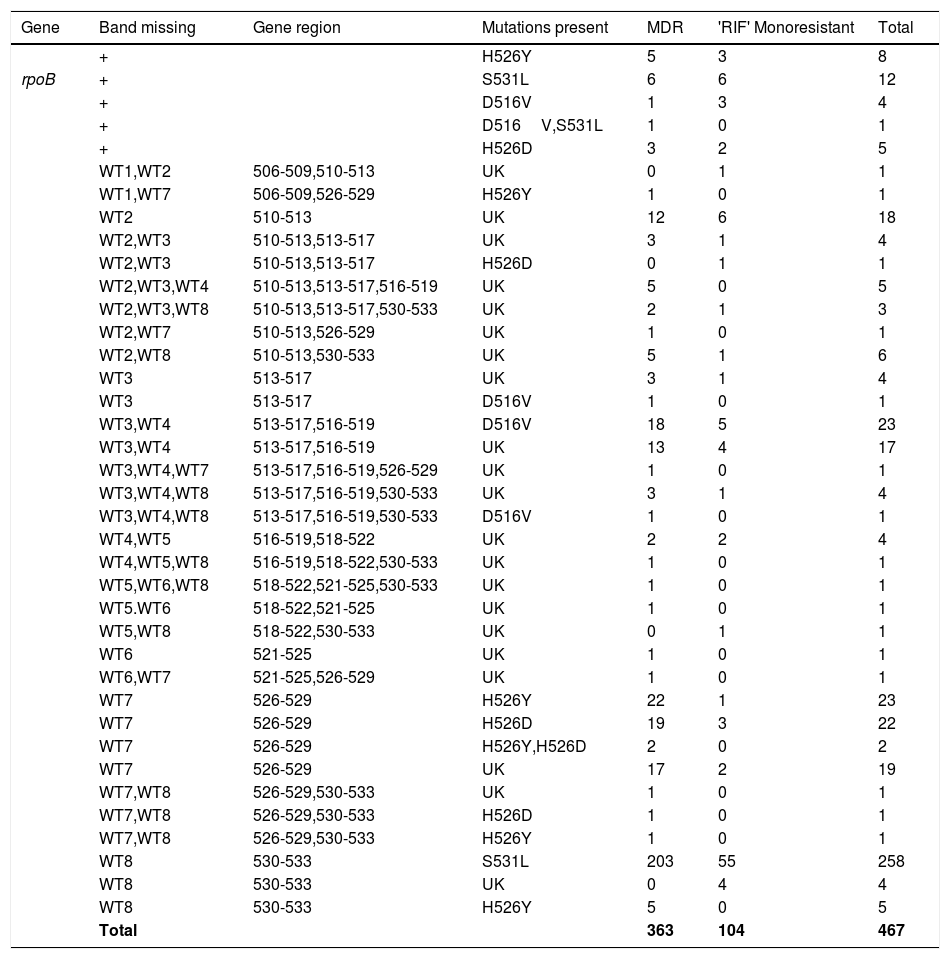
The Genotype MTBDR plus detected mutations, responsible for RIF and INH resistance, are displayed in Tables 2 and 3, respectively. Among all 467 RIF resistant isolates (363-MDR-TB strains and 104-RIF monoresistant), missing wild type (WT) band along with the presence of known mutant band was detected in 369 (79.01%) and missing WT with no gain in mutant band was found in 98 (20.98%) strains (i.e., no hybridization to the rpoB WT nor to either of the mutation probes). These isolates with the absence of both WT and mutant band were considered as unknown (UK). The most prominent known genetic mutation conferring RIF resistance was in codon S531L of rpoB gene (271/467; 58.8%) diagnosed by loss of the WT8 band and presence of MUT3 band, followed by H526Y mutation (40/467; 8.56%), D516V mutation (30/467; 6.42%), and H526D mutation (31/467; 6.63%). The mutation S531L was found in 210/363 (57.85%) and 61/104 (58.65%) of MDR strains and mono-RIF strains, respectively. This difference of rpoB S531L mutations in MDR-TB isolates compared with RIF monoresistant isolates was not statistically significant. In three MDR-TB strains, more than one mutation (D516V, S531L-(n=1) and H526Y, H526D-(n=2) was found in rpoB gene (Table 1) and were absent in RIF monoresistant isolates. Among these MDR-TB isolates with more than one mutation, one isolate showed mutations in two separate codons, that is, 516 and 531 and two isolates showed mutations in the same codon, that is, 526.
Gene mutation pattern as detected by GenoType MTBDRplus assay in Rifampicin resistant MTB isolates.
| Gene | Band missing | Gene region | Mutations present | MDR | 'RIF' Monoresistant | Total |
|---|---|---|---|---|---|---|
| + | H526Y | 5 | 3 | 8 | ||
| rpoB | + | S531L | 6 | 6 | 12 | |
| + | D516V | 1 | 3 | 4 | ||
| + | D516V,S531L | 1 | 0 | 1 | ||
| + | H526D | 3 | 2 | 5 | ||
| WT1,WT2 | 506-509,510-513 | UK | 0 | 1 | 1 | |
| WT1,WT7 | 506-509,526-529 | H526Y | 1 | 0 | 1 | |
| WT2 | 510-513 | UK | 12 | 6 | 18 | |
| WT2,WT3 | 510-513,513-517 | UK | 3 | 1 | 4 | |
| WT2,WT3 | 510-513,513-517 | H526D | 0 | 1 | 1 | |
| WT2,WT3,WT4 | 510-513,513-517,516-519 | UK | 5 | 0 | 5 | |
| WT2,WT3,WT8 | 510-513,513-517,530-533 | UK | 2 | 1 | 3 | |
| WT2,WT7 | 510-513,526-529 | UK | 1 | 0 | 1 | |
| WT2,WT8 | 510-513,530-533 | UK | 5 | 1 | 6 | |
| WT3 | 513-517 | UK | 3 | 1 | 4 | |
| WT3 | 513-517 | D516V | 1 | 0 | 1 | |
| WT3,WT4 | 513-517,516-519 | D516V | 18 | 5 | 23 | |
| WT3,WT4 | 513-517,516-519 | UK | 13 | 4 | 17 | |
| WT3,WT4,WT7 | 513-517,516-519,526-529 | UK | 1 | 0 | 1 | |
| WT3,WT4,WT8 | 513-517,516-519,530-533 | UK | 3 | 1 | 4 | |
| WT3,WT4,WT8 | 513-517,516-519,530-533 | D516V | 1 | 0 | 1 | |
| WT4,WT5 | 516-519,518-522 | UK | 2 | 2 | 4 | |
| WT4,WT5,WT8 | 516-519,518-522,530-533 | UK | 1 | 0 | 1 | |
| WT5,WT6,WT8 | 518-522,521-525,530-533 | UK | 1 | 0 | 1 | |
| WT5.WT6 | 518-522,521-525 | UK | 1 | 0 | 1 | |
| WT5,WT8 | 518-522,530-533 | UK | 0 | 1 | 1 | |
| WT6 | 521-525 | UK | 1 | 0 | 1 | |
| WT6,WT7 | 521-525,526-529 | UK | 1 | 0 | 1 | |
| WT7 | 526-529 | H526Y | 22 | 1 | 23 | |
| WT7 | 526-529 | H526D | 19 | 3 | 22 | |
| WT7 | 526-529 | H526Y,H526D | 2 | 0 | 2 | |
| WT7 | 526-529 | UK | 17 | 2 | 19 | |
| WT7,WT8 | 526-529,530-533 | UK | 1 | 0 | 1 | |
| WT7,WT8 | 526-529,530-533 | H526D | 1 | 0 | 1 | |
| WT7,WT8 | 526-529,530-533 | H526Y | 1 | 0 | 1 | |
| WT8 | 530-533 | S531L | 203 | 55 | 258 | |
| WT8 | 530-533 | UK | 0 | 4 | 4 | |
| WT8 | 530-533 | H526Y | 5 | 0 | 5 | |
| Total | 363 | 104 | 467 |
MDR, Multidrug resistant; RIF, Rifampicin; WT, Wild type; MUT, Mutant; UK, Unknown; +, Heteroresistant.
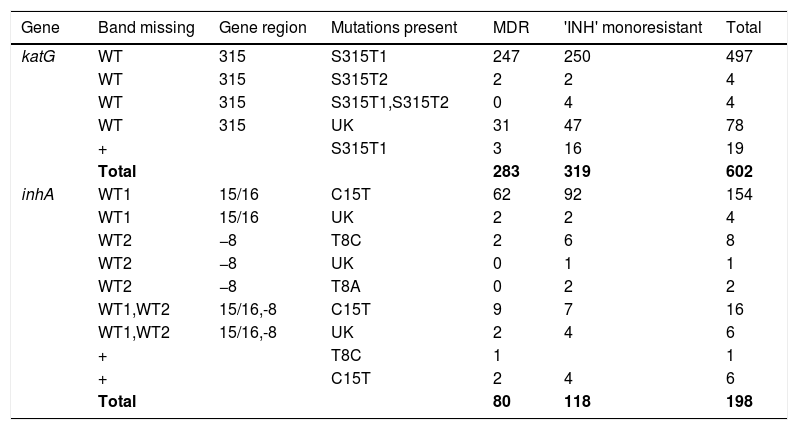
Pattern of gene mutations detected by GenoType MTBDRplus assay in Isoniazid drug-resistant MTB isolates.
| Gene | Band missing | Gene region | Mutations present | MDR | 'INH' monoresistant | Total |
|---|---|---|---|---|---|---|
| katG | WT | 315 | S315T1 | 247 | 250 | 497 |
| WT | 315 | S315T2 | 2 | 2 | 4 | |
| WT | 315 | S315T1,S315T2 | 0 | 4 | 4 | |
| WT | 315 | UK | 31 | 47 | 78 | |
| + | S315T1 | 3 | 16 | 19 | ||
| Total | 283 | 319 | 602 | |||
| inhA | WT1 | 15/16 | C15T | 62 | 92 | 154 |
| WT1 | 15/16 | UK | 2 | 2 | 4 | |
| WT2 | −8 | T8C | 2 | 6 | 8 | |
| WT2 | −8 | UK | 0 | 1 | 1 | |
| WT2 | −8 | T8A | 0 | 2 | 2 | |
| WT1,WT2 | 15/16,-8 | C15T | 9 | 7 | 16 | |
| WT1,WT2 | 15/16,-8 | UK | 2 | 4 | 6 | |
| + | T8C | 1 | 1 | |||
| + | C15T | 2 | 4 | 6 | ||
| Total | 80 | 118 | 198 |
MDR, Multidrug Resistant; INH, Isoniazid; WT, Wild type; UK, Unknown; +, Heteroresistant.
Out of 98 (20.98%) RIF-resistant isolates with unknown mutations or mutant bands, 25 were observed in RIF monoresistant strains and 73 were observed in MDR isolates. The 98 isolates included missing WT1/WT2 (1; 1.02%), WT2 (18; 18.36%), WT2/WT3 (4; 4.08%), WT2/WT3/WT4 (5; 5.10%), WT2/WT3/WT8 (3; 3.80%), WT2/WT7 (1; 1.02%) WT2/WT8 (6; 6.12%), WT3 (4; 4.08%), WT3/WT4 (17; 17.34%), WT3/WT4/WT7 (1; 1.02%), WT3/WT4/WT8 (4; 4.08%), WT4/WT5 (4; 4.08%), WT4/WT5/WT8 (1; 1.02%), WT5/WT6/WT8 (1; 1.02%), WT5/WT6 (1; 1.02%), WT5/WT8 (1; 1.02%), WT6 (1; 1.02%), WT6/WT7 (1; 1.02%), WT7(19; 19.38%), WT7/WT8 (1; 1.02%), WT8 (4; 4.08%). Mixed pattern to RIF with all WT probes present along with the presence of one or more additional mutant bands was found in 30/467 (6.42%), most common being S531L (13/30; 43.3%), followed by H526Y (8/30; 26.6%), D516V (5/30; 16.6%), and H526D (5/30; 16.6%), while one MDR-TB isolate with D516V had an additional mutation, S531L in the rpoB gene.
Out of 800 INH-resistant isolates detected by GenoType MTBDRplus, mutations in katG were found in 602/800 (75.25%) isolates and mutations in inhA were found in 198 (24.75%) isolates. Out of 602 INH-resistant isolates, mutations in katG were found in 319 (53%) INH-monoresistant isolates and in 283 (47%) MDR isolates. Known mutations were detected at S315T1 (MUT1 band) in 520/602 (86.37%) and S315T2 in 8/602 (1.32%) of the katG mutant isolates. Four INH-mono resistant isolates showed the presence of both S315T1 and S315T2 mutations in katG and were absent in MDR-TB isolates. Missing WTs with the absence of mutant band were found in 78 (12.95%) of katG mutants, and out of these, 31 were found in MDR isolates and 47 in INH katG-monoresistant strains. Mixed pattern to INH with all WT probes present along with the presence of one or more mutant bands was found in 19 (3.15%) isolates, known mutation, S315T1 was present in all the isolates of both MDR (3/19) and INH katG monoresistant isolates (16/19).
Mutations in inhA were found in 198 (24.75%) INH-resistant isolates, of which 118 were in inhA INH-monoresistant isolates and 80 were in MDR isolates. The most commonly known genetic mutation observed in inhA regulatory region was C15T (176/198; 88.88%), followed by T8C (9/198; 4.54%) and T8A (2/198; 1.00%). Missing WTs with unknown mutations were found in 11 (5.55%) inhA mutant isolates. Mixed WT and mutant pattern to INH was found in 7/198 (3.5%) of the total INH-resistant isolates. 15/437 (3.4%) of the INH monoresistant isolates had mutations in both inhA and katG genes. Mutations in C15T were relatively higher in INH mono-resistant (103/118) than MDR strains (73/80), but the difference was not significant. Mutations in rpoB and katG gene account for 78% (283/363) of all MDR cases and mutations in rpoB and inhA genes account for 22% (80/363) of all the MDR cases. No MDR case was found having mutations in all three genes (rpoB, katG & inhA).
DiscussionAndhra Pradesh (AP) is one of the largest southern states in India and it records 1, 07,293 patients, annually, enrolled under RNTCP program for TB treatment with ca. 8% of the suspected harboring MDR TB (http://www.tbfacts.org/tb-statistics-india/, 2015). Since 2006, AP has been implementing the second line anti-TB treatment under the Directly Observed Treatment Short course plus program (DOTSplus) of RNTCP for diagnosis and treatment. In this study, we aimed to determine the patterns of drug resistance against two first line anti-TB drugs (RIF and INH). We also analyzed the frequency and type of drug resistant mutations in the 81-bp hotspot region of rpoB gene, codon 315 of katG gene and alterations in the inhA promoter region found upstream from the mabA-inhA locus in clinical isolates from pulmonary TB patients of southern districts of AP, India, enrolled under the RNTCP program using the commercially available GenoType® MTBDRplus assay.
The overall RIF resistance in our study was 51.65% (467/904) of drug resistant isolates and is considered as surrogate marker for detection of MDR TB. The isolates monoresistant to RIF were also considered under MDR category and the patients were given treatment under category IV of DOTSplus program (treatment given to MDR patients). Similar findings were reported by Janmeja and Raj, 1998, from Haryana (49%)12 and Rawat et al., 2009, from Uttarakhand (57.22%).13 However, percent RIF resistance observed in our study was much lesser than the percent RIF resistance reported in other studies by Singhalet al., 2015 from New Delhi, India (73.9%)14 and Barnard et al., 2008 from South Africa (91%).15 Among RIF-resistant MTB isolates, 58.8% had a single nucleotide Ser-to-Leu substitution at rpoB 531 position followed by His-to-Asp at 526, Asp-to-Val at 516, and His-to-Tyr at 526 positions. These data are in accordance with the mutation frequencies reported previously by other studies.8
According to the results of many earlier studies, the most commonly observed mutations among all the mutations leading to RIF resistance was in the 531 region of rpoB gene, mainly S531L missense mutation (58.8%). Most of the point mutations observed in this study were in the codons 531, 516, and 526. These results coincide with the previous results reported by others. The frequency of the major mutation S531L (serine to threonine substitution) (58.8%) observed in our study is consistent with those found in other studies in India ((Singhal et al., 2015 (59.0%),14 Lingala et al., 2010 (53.6%),16 Maurya et al., 2013 (62.3%),17 and Raizadaet al., 2014 (47%)9), Nepal (Sharma et al., 2014 (50%), and South Vietnam (Huyen et al., 2010 (50%)).18,19 Nonetheless, the frequency of S531L was found to be lower than some of the other reports from India (Yadav et al., 2013 (72%), Mohan et al., 2014 (81.5%) and Raveendran et al., 2012 (84.6%)), Germany (Hillemann et al., 2007 (73.6%)), Denmark (Vijdea et al., (86%)), and Russia (Nikolayevskyy et al., 2010 (94%)).6,20–24 In the present study, frequency of S531L mutation was nearly same in both RIF monoresistant isolates (55/104; 53%) and MDR-TB isolates (203/363; 56%). This is in contrast to other reports from India and South Africa, where the S531L mutation was significantly higher in MDR-TB than in RIF monoresistant strains.14,15,25 However, these results were in concordance with the results of Yadav et al., 2013 and Raizadaet al., 2014.6,9
For INH resistance, it has been reported previously by many authors that the most common mutation observed was in the 315 region of katG, which was present in 75.25% (602/800) of all INH resistant isolates and the S315T1 mutation (497/602; 82.5%) that led amino acid serine to threonine substitution has also been related to high level of resistance to INH. These are in accordance with the worldwide reported figures (50–90%) of all observed phenotypic INH resistance associated with the katG315 mutation. In TB endemic countries, it has been reported that high prevalence of katG mutation was associated with a majority of the INH resistant isolates.17,19,26–28
Mutations in inhA promoter were found in 24.75% (198/800) INH resistance, while C15T being the most common mutation (85.85%; 170/198). The inhA mutations accounted for 27% (80/363) and 20.88% (118/437) of mono-INH and MDR strains, respectively. Out of 198 total INH drug resistant mutations, 60% (118/198) of inhA mutations were found in INH monoresistant and 40% (80/118) were in MDR TB isolates. Mutation occurred in 24.75% of INH resistant MTB strains in the regulatory region of inhA (invariably C15T), which is similar to other studies by Singhal et al., 2015 (13.4%), Huyen et al., 2010 (18%), and Brossier et al., 2006 (17%), but considerably lower than that reported by Barnard et al., 2008 (40%).8,14,19 Low or intermediate levels of INH resistance was shown to be associated with the mutations of regulatory region of inhA gene (20–35%) by various studies, which are in agreement with our results.29
In our study we did not find any combined mutations in katG and inhA, as compared with other studies.6,30 Unlike RIF resistance that has been shown to be a surrogate marker for detection of MDR TB, the clinical implication of the low level INH resistance needs further investigations. We did not find any association between a particular mutation in katG and/or inhA and the occurrence of INH monoresistance or MDR TB. In contrast to our results, previous studies by Barnard et al., 2008, and Bolotin et al., 2009, reported a significantly higher level of association of S315T of katG and/or inhA mutations in MDR-TB isolates than in INH monoresistant TB isolates. Mutations in the katG gene of strains resistant to INH were observed at higher frequency compared with the inhA promoter mutations.15,31 Unlike RIF resistance mutations (specific to 81-bp region of the rpoB gene), mutations causing resistance to INH are more complex and involves mutations in several genes such as katG, ahpC, fabG1, kasA, furA, ndh and oxyR-ahpC intergenic region.32,33 Considering this, LPA test might have failed to detect all mutations outside the katG gene that is, in other genomic regions that confer INH resistance in some of the drug resistant isolates. Further sequencing of TB strains would benefit to characterize the mutations in the gene region other than katG gene and inhA promoter.
Overall MDR-TB rate observed in this study was 12.54% (363), which is slightly lower than that in other studies reported from other parts of India (Chandigarh (27.6%),34 Tamil Nadu (25%),35 Andhra Pradesh (26.7%),20 and Gujarat (30.2%)36). In contrast, higher rates of multidrug resistance were observed in Dehradun (57.22%)13 and Delhi 53.6%37 and rates are in concordance with those in Sewagram Wardha (9.2–9.6%).38
In this study, we observed significantly higher number of isolates with missing WT band and without mutant band and were in the rate of 21% (n=98/467) in rpoB, 13% (n=78/602) in katG, and 5.55% (n=11/198) in inhA, which indicate that the mutations conferring drug resistance are not the common mutations of the genic region that are incorporated in the present MDRTB plus strip and are probably rare mutations. Interestingly, such unknown mutations were found comparatively more in RIF monoresistant (24.03% vs 20%) and INH monoresistant isolates (12.5% vs 9.6%) than in MDR-TB isolates. But the difference observed was not statistically significant. Similarly, higher number of new mutations have been reported from India,14,39 Vietnam19 and Uganda.40 Further characterization of these gene mutations is necessary in order to detect new and emerging drug resistance mutations. It would be interesting to know any direct relationship between specific mutations detected with treatment outcomes of INH monoresistant cases.
Another interesting observation was a higher percent of TB isolates from female patients that were found to be MDR (100/579; 17.27%) compared to male patients (263/2481; 10.56%), though the majority of the isolates were from male patients (2481/3060, 81.10%) compared to female patients (579/3060, 18.92%). The difference between the two was statistically significant (χ2=21.863, p<0.005). These findings were in line with other studies reported by Bazira et al., 2010 and Lomtadze et al., 2009. This might be related to health seeking behavior, with prolonged delays in female patients (probably due to lack of control of financial resources at household levels) as has been reported by Bazira et al., 2010.41,42 However, reasons for the increased risk of MDR TB among women patients observed in our region are not known and require further investigation.
Patients’ age-wise distribution of MDR TB also revealed some interesting observations. Of 2894 MTB positive patients, 86% of the patients were in the age group of 20–50 years. Intriguingly, high percent of MDR-TB (29/363; 18.12%) cases were reported in the age group of below 20 years. Of the 363 MDR-TB isolates, 187/363 (13.26%) were from the age group 21–40 years and 125/363(9.78%) were from the age group 41–60 years (χ2=17.43, p<0.005). As young adult males are from economically productive section of society, high MDR-TB burden in them have many socio-economic implications. We could not get the information regarding other reported clinical and socio-demographic risk factors such as alcoholism, smoking, number of previous treatments, irregular treatments, lung cavities in chest X-ray, occupation, educational status, monthly income, site of TB disease, and diabetes. Further studies are warranted to analyze the risk factors of MDR TB in this region.
In India, MDR TB among notified new pulmonary TB patients is 2.2% with 95%CI of 1.9–2.6% and MDR-TB among previously treated pulmonary TB cases is 15% with 95%CI of 11–19%.43 Globally, 3.7% of new cases and 20% of previously treated cases are estimated to be MDR TB. In the present study, the prevalence of MDR TB cases was high in previously treated cases (12.21%; 344/2817) and relatively low in new cases (7.81%; 19/243). The difference between the two was statistically significant (χ2=6.4, p<0.05). The results of the present study are in concordance with the figures reported by RNTCP and WHO global surveys. Gupta et al., 2014, and Kauret al., 2016, had also observed the significantly higher number of MDR TB in previously treated cases compared to new cases.44,45 As found in many other anti-TB drug resistance surveillance studies, our study also suggests that history of anti-tubercular treatment has been consistently associated with the risk of MDR TB.14,44–46 Similarly, primary monoresistance to RIF was observed in five (2.05%) cases and acquired resistance in 99 (3.51%) cases. Primary INH monoresistance was detected in 29 (11.93%) cases and acquired resistance in 408 (14.48%) cases.
TB is the most common opportunistic bacterial infection among the human immunodeficiency syndrome (HIV) infected patients. Furthermore, MDR-TB co-infected HIV patients are at higher risk of death than the HIV non-infected individuals in developing countries like India.47 In our study, out of 2894 TB positive patients, 227 patients (7.41%) were found to be co-infected with HIV. Out of 227 HIV co-infected patient samples, 184 (81.05%) were drug sensitive, 25 (11.01%) were INH resistant, 4 (1.76%) were RIF resistant, and 14 (6.16%) were MDR (resistant to both INH and RIF). MDR was seen in only 6.16% isolates as compared to 12.31% of isolates from HIV negative cases (χ2=7.114, p<0.05). Lower incidence of MDR TB in HIV-infected patients has also been reported by Praharaj et al., 2004. However, the rates of HIV/TB co-infection in different regions of India were reported to be between 0.4% and 30%.48,49
Another important observation is presence of hetero-resistant population characterized by presence of both WT and the mutant probes corresponding to sensitive or susceptible and resistant isolate, respectively. High rate of hetero-resistance pattern was observed in all three genes, that is, rpoB (6.42%; 30/467), katG (3.15%; 19/602), and inhA promoter region (3.5%; (7/198). The most frequent mutation was S531L (40.0%) in rpoB region, S315T1 (100%) in katG gene, and C15T in inhA promoter region (85.7%). In our study, comparatively, hetero-resistant population was more in RIF monoresistant and INH monoresistant isolates than in MDR-TB cases. A similar report of high prevalence of hetero-resistant TB isolates were reported in other studies by Singh et al., 2014, Tolani et al., 2012, and ÇAVUŞOĞLU et al., 2011.30 The presence of hetero-resistant TB isolates indicates the slow evolution of mycobacteria from a sensitive to resistant profile during drug treatment and future upsurge of MDR TB in this region. It also reflects the super infection of both sensitive and resistant isolates in the same patient who must be given a drug resistant TB regime. The studied risk factors such as age, sex, history of TB, and HIV status were not significantly associated with the observed hetero-resistant (data not shown). Detection of hetero-resistant TB isolates is another advantage for the use of LPA.
Majority of LPA invalid results of the present study were found in culture negative samples or sputum specimens with lower bacillary load (1+) and their frequency inversely correlated to smear grading as has been reported by previous studies.15,39 The results of the present study also support the unsuitability of LPA test for direct use on smear-negative clinical specimens and also low specificity of the smear microscopy, which has been used as a standard point of care test to diagnose MTB.6,50 Further characterization of smear-positive and LPA-invalid samples using phenotypic and biochemical assays suggested the prevalence of NTM (n=15) in this region. Using these assays, we could identify NTM up to genus level only and further studies are warranted to identify NTM species spectra, so as to initiate appropriate treatment to further curb the spread of NTM. The results of the present study also emphasize the need to isolate the causative organism in pure culture, as smear microscopy does not distinguish MTBC that causes pulmonary TB from NTMs. Surveillance or epidemiological studies should be done further to control spread of these infections.
Though the GenoType MTBDRplus has high accuracy coupled with reduced turnaround time, the test can only detect drug resistance mutations that originate from rpoB, katG, and inhA. The assay cannot detect mutations originating from other genomic regions and other resistance mechanisms. This is especially true in detecting INH resistance and explains the comparatively low sensitivity of the assay in detecting INH resistance. Secondly, high possibility of silent mutations, that is, mutations that do not lead to change in amino acid and hence the strain being sensitive in phenotypic drug susceptibility studies. The reported turnaround testing time including specimen transportation time was slightly higher (five days) than the RNTCP recommended TAT, that is, within five days. Theoretically, LPA was completed within three days, however, the observed TAT, that is, five days, was mainly due to lag in transportation of samples from primary health center to the testing laboratory. The patients who were diagnosed with MDR TB were provided with category IV treatment of DOTS plus program within 10 days of test results.
In conclusion, the present study ascertained the prevalence of drug-resistant TB isolates in southern districts of Andhra Pradesh and also the pattern of rpoB, katG, and inhA gene mutations in the drug-resistant isolates. This study revealed that the presence of MDR TB is a major serious public health problem of this region. Further studies are warranted to know the transmission dynamics of these MDR TB isolates so as to curb further community and nosocomial transmission of drug resistant TB, especially, limiting MDR-TB and XDR-TB disease progression. Our study also reported a number of unknown or uncommon mutations prevalent in this region, which are not covered by routinely used molecular diagnostic kits. Specific mutations found by the MTBDRplus assay may help in empiric choice of an anti-TB treatment regimen. Continued surveillance through traditional culture and DST methods will remain important to individualize and customize treatment regimens for drug-resistant MTB.
FundingThe consumables and machines for GenoType MTBDRplus assay have been provided free of cost by Damien Foundation India Trust, India, for management of MDR-TB patients under the programmatic management of Drug resistant TB (PMDT).
Ethical approvalThis study received ethical clearance from the ethics committee of Krishna Institute of Medical Sciences (KIMS), Nellore.
Informed consentVerbal informed consent was obtained from all participants included in this study.
Conflict of interestThe researchers claim No conflicts of interest.
We acknowledge the technical help of administrative and laboratory staff of Damien TB research centre, Nellore. We also acknowledge the support extended by DTCO’S of southern districts of Andhra Pradesh. We also thank Debasmita N, Gopala Krishna M and Sangeetha Chakrabortty for critical appraisal of the article. Dr. Uday Sankar Allam acknowledges Department of Science and Technology (DST) for their support through Early Career Research Award (ECRA). Mohammad Shaik Jasmine acknowledges University Grants Commission (UGC) for Maulana Azad National Fellowship.